A peek into tropomyosin binding and unfolding on the actin filament
- PMID: 19629180
- PMCID: PMC2710508
- DOI: 10.1371/journal.pone.0006336
A peek into tropomyosin binding and unfolding on the actin filament
Abstract
Background: Tropomyosin is a prototypical coiled coil along its length with subtle variations in structure that allow interactions with actin and other proteins. Actin binding globally stabilizes tropomyosin. Tropomyosin-actin interaction occurs periodically along the length of tropomyosin. However, it is not well understood how tropomyosin binds actin.
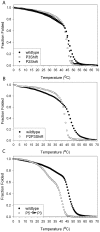
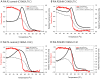
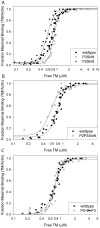
Principal findings: Tropomyosin's periodic binding sites make differential contributions to two components of actin binding, cooperativity and affinity, and can be classified as primary or secondary sites. We show through mutagenesis and analysis of recombinant striated muscle alpha-tropomyosins that primary actin binding sites have a destabilizing coiled-coil interface, typically alanine-rich, embedded within a non-interface recognition sequence. Introduction of an Ala cluster in place of the native, more stable interface in period 2 and/or period 3 sites (of seven) increased the affinity or cooperativity of actin binding, analysed by cosedimentation and differential scanning calorimetry. Replacement of period 3 with period 5 sequence, an unstable region of known importance for cooperative actin binding, increased the cooperativity of binding. Introduction of the fluorescent probe, pyrene, near the mutation sites in periods 2 and 3 reported local instability, stabilization by actin binding, and local unfolding before or coincident with dissociation from actin (measured using light scattering), and chain dissociation (analyzed using circular dichroism).
Conclusions: This, and previous work, suggests that regions of tropomyosin involved in binding actin have non-interface residues specific for interaction with actin and an unstable interface that is locally stabilized upon binding. The destabilized interface allows residues on the coiled-coil surface to obtain an optimal conformation for interaction with actin by increasing the number of local substates that the side chains can sample. We suggest that local disorder is a property typical of coiled coil binding sites and proteins that have multiple binding partners, of which tropomyosin is one type.
Conflict of interest statement
Figures


Similar articles
-
Tropomyosin's periods are quasi-equivalent for actin binding but have specific regulatory functions.Biochemistry. 2007 Dec 25;46(51):14917-27. doi: 10.1021/bi701570b. Epub 2007 Dec 4. Biochemistry. 2007. PMID: 18052203
-
Local destabilization of the tropomyosin coiled coil gives the molecular flexibility required for actin binding.Biochemistry. 2003 Dec 9;42(48):14114-21. doi: 10.1021/bi0348462. Biochemistry. 2003. PMID: 14640678
-
Dual requirement for flexibility and specificity for binding of the coiled-coil tropomyosin to its target, actin.Structure. 2006 Jan;14(1):43-50. doi: 10.1016/j.str.2005.09.016. Structure. 2006. PMID: 16407064
-
Periodicities designed in the tropomyosin sequence and structure define its functions.Bioarchitecture. 2013 May-Jun;3(3):51-6. doi: 10.4161/bioa.25616. Epub 2013 Jul 8. Bioarchitecture. 2013. PMID: 23887197 Free PMC article. Review.
-
Tropomyosin: function follows structure.Adv Exp Med Biol. 2008;644:60-72. doi: 10.1007/978-0-387-85766-4_5. Adv Exp Med Biol. 2008. PMID: 19209813 Review.
Cited by
-
Pyrene: a probe to study protein conformation and conformational changes.Molecules. 2011 Sep 14;16(9):7909-35. doi: 10.3390/molecules16097909. Molecules. 2011. PMID: 22143550 Free PMC article. Review.
-
A periodic pattern of evolutionarily conserved basic and acidic residues constitutes the binding interface of actin-tropomyosin.J Biol Chem. 2013 Apr 5;288(14):9602-9609. doi: 10.1074/jbc.M113.451161. Epub 2013 Feb 18. J Biol Chem. 2013. PMID: 23420843 Free PMC article.
-
Investigations into tropomyosin function using mouse models.J Mol Cell Cardiol. 2010 May;48(5):893-8. doi: 10.1016/j.yjmcc.2009.10.003. Epub 2009 Oct 14. J Mol Cell Cardiol. 2010. PMID: 19835881 Free PMC article. Review.
-
Instability in the central region of tropomyosin modulates the function of its overlapping ends.Biophys J. 2013 Nov 5;105(9):2104-13. doi: 10.1016/j.bpj.2013.09.026. Biophys J. 2013. PMID: 24209855 Free PMC article.
-
Evolutionarily conserved surface residues constitute actin binding sites of tropomyosin.Proc Natl Acad Sci U S A. 2011 Jun 21;108(25):10150-5. doi: 10.1073/pnas.1101221108. Epub 2011 Jun 3. Proc Natl Acad Sci U S A. 2011. PMID: 21642532 Free PMC article.
References
-
- Burkhard P, Stetefeld J, Strelkov SV. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 2001;11:82–88. - PubMed
-
- Crick FHC. The packing of alpha-helices. Simple coiled-coils. Acta Crystallographica. 1953;6:689–697.
-
- Gunning PW, Schevzov G, Kee AJ, Hardeman EC. Tropomyosin isoforms: divining rods for actin cytoskeleton function. Trends Cell Biol. 2005;15:333–341. - PubMed
-
- Perry SV. Vertebrate tropomyosin: distribution, properties and function. J Muscle Res Cell Motil. 2001;22:5–49. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

