Isothermal DNA amplification in vitro: the helicase-dependent amplification system
- PMID: 19629390
- PMCID: PMC11115679
- DOI: 10.1007/s00018-009-0094-3
Isothermal DNA amplification in vitro: the helicase-dependent amplification system
Abstract
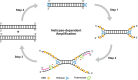
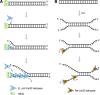
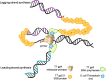
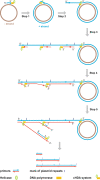
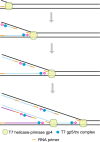
Since the development of polymerase chain reaction, amplification of nucleic acids has emerged as an elemental tool for molecular biology, genomics, and biotechnology. Amplification methods often use temperature cycling to exponentially amplify nucleic acids; however, isothermal amplification methods have also been developed, which do not require heating the double-stranded nucleic acid to dissociate the synthesized products from templates. Among the several methods used for isothermal DNA amplification, the helicase-dependent amplification (HDA) is discussed in this review with an emphasis on the reconstituted DNA replication system. Since DNA helicase can unwind the double-stranded DNA without the need for heating, the HDA system provides a very useful tool to amplify DNA in vitro under isothermal conditions with a simplified reaction scheme. This review describes components and detailed aspects of current HDA systems using Escherichia coli UvrD helicase and T7 bacteriophage gp4 helicase with consideration of the processivity and efficiency of DNA amplification.
Figures
Similar articles
-
Characterization of a thermostable UvrD helicase and its participation in helicase-dependent amplification.J Biol Chem. 2005 Aug 12;280(32):28952-8. doi: 10.1074/jbc.M503096200. Epub 2005 Jun 13. J Biol Chem. 2005. PMID: 15955821 Free PMC article.
-
Simultaneous amplification and screening of whole plasmids using the T7 bacteriophage replisome.Nucleic Acids Res. 2006 Aug 7;34(13):e98. doi: 10.1093/nar/gkl547. Nucleic Acids Res. 2006. PMID: 16893951 Free PMC article.
-
Helicase-dependent isothermal DNA amplification.EMBO Rep. 2004 Aug;5(8):795-800. doi: 10.1038/sj.embor.7400200. Epub 2004 Jul 9. EMBO Rep. 2004. PMID: 15247927 Free PMC article.
-
DNA Helicase-Polymerase Coupling in Bacteriophage DNA Replication.Viruses. 2021 Aug 31;13(9):1739. doi: 10.3390/v13091739. Viruses. 2021. PMID: 34578319 Free PMC article. Review.
-
Mechanisms of a ring shaped helicase.Nucleic Acids Res. 2006;34(15):4216-24. doi: 10.1093/nar/gkl508. Epub 2006 Aug 25. Nucleic Acids Res. 2006. PMID: 16935879 Free PMC article. Review.
Cited by
-
Recent progress in diagnosis and treatment of Human African Trypanosomiasis has made the elimination of this disease a realistic target by 2030.Front Med (Lausanne). 2022 Nov 3;9:1037094. doi: 10.3389/fmed.2022.1037094. eCollection 2022. Front Med (Lausanne). 2022. PMID: 36405602 Free PMC article. Review.
-
Nucleic acid testing for tuberculosis at the point-of-care in high-burden countries.Expert Rev Mol Diagn. 2012 Sep;12(7):687-701. doi: 10.1586/erm.12.71. Expert Rev Mol Diagn. 2012. PMID: 23153237 Free PMC article. Review.
-
HCV Detection, Discrimination, and Genotyping Technologies.Sensors (Basel). 2018 Oct 12;18(10):3423. doi: 10.3390/s18103423. Sensors (Basel). 2018. PMID: 30322029 Free PMC article. Review.
-
Prospects of Microfluidic Technology in Nucleic Acid Detection Approaches.Biosensors (Basel). 2023 May 27;13(6):584. doi: 10.3390/bios13060584. Biosensors (Basel). 2023. PMID: 37366949 Free PMC article. Review.
-
Alteration of enzymes and their application to nucleic acid amplification (Review).Int J Mol Med. 2020 Nov;46(5):1633-1643. doi: 10.3892/ijmm.2020.4726. Epub 2020 Sep 15. Int J Mol Med. 2020. PMID: 33000189 Free PMC article. Review.
References
-
- Mullis KB. The unusual origin of the polymerase chain reaction. Sci Am. 1990;262:56–61. - PubMed
-
- Mullis KB. Target amplification for DNA analysis by the polymerase chain reaction. Ann Biol Clin (Paris) 1990;48:579–582. - PubMed
-
- Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;24:476–480. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

