Highly conserved histidine plays a dual catalytic role in protein splicing: a pKa shift mechanism
- PMID: 19630416
- PMCID: PMC2737186
- DOI: 10.1021/ja904318w
Highly conserved histidine plays a dual catalytic role in protein splicing: a pKa shift mechanism
Abstract
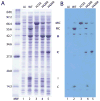
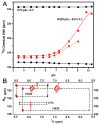
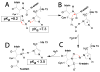
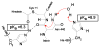
Protein splicing is a precise autocatalytic process in which an intein excises itself from a precursor with the concomitant ligation of the flanking sequences. Protein splicing occurs through acid-base catalysis in which the ionization states of active site residues are crucial to the reaction mechanism. In inteins, several conserved histidines have been shown to play important roles in protein splicing, including the most conserved "B-block" histidine. In this study, we have combined NMR pK(a) determination with quantum mechanics/molecular mechanics (QM/MM) modeling to study engineered inteins from Mycobacterium tuberculosis (Mtu) RecA intein. We demonstrate a dramatic pK(a) shift for the invariant B-block histidine, the most conserved residue among inteins. The B-block histidine has a pK(a) of 7.3 +/- 0.6 in a precursor and a pK(a) of <3.5 in a spliced intein. The pK(a) values and QM/MM data suggest that the B-block histidine has a dual role in the acid-base catalysis of protein splicing. This histidine likely acts as a general base to initiate splicing with an acyl shift and then as a general acid to cause the breakdown of the scissile bond at the N-terminal splicing junction. The proposed pK(a) shift mechanism accounts for the biochemical data supporting the essential role for the B-block histidine and for the near absolute sequence conservation of this residue.
Figures


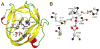

Similar articles
-
Unveiling the Catalytic Role of B-Block Histidine in the N-S Acyl Shift Step of Protein Splicing.J Phys Chem B. 2017 Aug 24;121(33):7786-7796. doi: 10.1021/acs.jpcb.7b04276. Epub 2017 Aug 11. J Phys Chem B. 2017. PMID: 28737941
-
Crystal structures of an intein from the split dnaE gene of Synechocystis sp. PCC6803 reveal the catalytic model without the penultimate histidine and the mechanism of zinc ion inhibition of protein splicing.J Mol Biol. 2005 Nov 11;353(5):1093-105. doi: 10.1016/j.jmb.2005.09.039. Epub 2005 Sep 30. J Mol Biol. 2005. PMID: 16219320
-
pK(a) coupling at the intein active site: implications for the coordination mechanism of protein splicing with a conserved aspartate.J Am Chem Soc. 2011 Jul 6;133(26):10275-82. doi: 10.1021/ja203209f. Epub 2011 Jun 9. J Am Chem Soc. 2011. PMID: 21604815 Free PMC article.
-
Protein splicing in cis and in trans.Chem Rec. 2006;6(4):183-93. doi: 10.1002/tcr.20082. Chem Rec. 2006. PMID: 16900466 Review.
-
Inteins, valuable genetic elements in molecular biology and biotechnology.Appl Microbiol Biotechnol. 2010 Jun;87(2):479-89. doi: 10.1007/s00253-010-2628-x. Epub 2010 May 7. Appl Microbiol Biotechnol. 2010. PMID: 20449740 Free PMC article. Review.
Cited by
-
Protein splicing: how inteins escape from precursor proteins.J Biol Chem. 2014 May 23;289(21):14498-505. doi: 10.1074/jbc.R113.540310. Epub 2014 Apr 2. J Biol Chem. 2014. PMID: 24695729 Free PMC article. Review.
-
¹H, ¹³C and ¹⁵N NMR assignments of a Drosophila Hedgehog autoprocessing domain.Biomol NMR Assign. 2014 Oct;8(2):279-81. doi: 10.1007/s12104-013-9500-8. Epub 2013 Jun 14. Biomol NMR Assign. 2014. PMID: 23765287 Free PMC article.
-
Mutual synergistic protein folding in split intein.Biosci Rep. 2012 Oct;32(5):433-42. doi: 10.1042/BSR20120049. Biosci Rep. 2012. PMID: 22681309 Free PMC article.
-
A Conserved Histidine Residue Drives Extein Dependence in an Enhanced Atypically Split Intein.J Am Chem Soc. 2022 Oct 19;144(41):19196-19203. doi: 10.1021/jacs.2c08985. Epub 2022 Oct 4. J Am Chem Soc. 2022. PMID: 36194550 Free PMC article.
-
1H, 13C, and 15N NMR assignments of the Pyrococcus abyssi DNA polymerase II intein.Biomol NMR Assign. 2011 Oct;5(2):233-5. doi: 10.1007/s12104-011-9307-4. Epub 2011 Apr 26. Biomol NMR Assign. 2011. PMID: 21519863 Free PMC article.
References
-
- Paulus H. Chemical Society Reviews. 1998;27:375–386.
-
- Belfort M. In: Homing Endonucleases and Inteins. Belfort M, Derbyshire V, Stoddard BL, Wood DW, editors. Vol. 16. Springer; Berlin Heidelberg: 2005. pp. 1–9.
-
- Blaschke UK, Silberstein J, Muir TW. Methods Enzymol. 2000;328:478–96. - PubMed
-
- Mootz HD, Blum ES, Tyszkiewicz AB, Muir TW. J Am Chem Soc. 2003;125:10561–9. - PubMed
-
- Tyszkiewicz AB, Muir TW. Nat Methods. 2008 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

