Rad1, rad10 and rad52 mutations reduce the increase of microhomology length during radiation-induced microhomology-mediated illegitimate recombination in saccharomyces cerevisiae
- PMID: 19630519
- PMCID: PMC2741414
- DOI: 10.1667/RR1675.1
Rad1, rad10 and rad52 mutations reduce the increase of microhomology length during radiation-induced microhomology-mediated illegitimate recombination in saccharomyces cerevisiae
Abstract
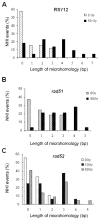
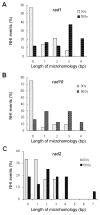
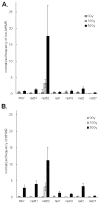
Abstract Illegitimate recombination can repair DNA double-strand breaks in one of two ways, either without sequence homology or by using a few base pairs of homology at the junctions. The second process is known as microhomology-mediated recombination. Previous studies showed that ionizing radiation and restriction enzymes increase the frequency of microhomology-mediated recombination in trans during rejoining of unirradiated plasmids or during integration of plasmids into the genome. Here we show that radiation-induced microhomology-mediated recombination is reduced by deletion of RAD52, RAD1 and RAD10 but is not affected by deletion of RAD51 and RAD2. The rad52 mutant did not change the frequency of radiation-induced microhomology-mediated recombination but rather reduced the length of microhomology required to undergo repair during radiation-induced recombination. The rad1 and rad10 mutants exhibited a smaller increase in the frequency of radiation-induced microhomology-mediated recombination, and the radiation-induced integration junctions from these mutants did not show more than 4 bp of microhomology. These results suggest that Rad52 facilitates annealing of short homologous sequences during integration and that Rad1/Rad10 endonuclease mediates removal of the displaced 3' single-stranded DNA ends after base-pairing of microhomology sequences, when more than 4 bp of microhomology are used. Taken together, these results suggest that radiation-induced microhomology-mediated recombination is under the same genetic control as the single-strand annealing apparatus that requires the RAD52, RAD1 and RAD10 genes.
Figures





Similar articles
-
Microarray-based genetic screen defines SAW1, a gene required for Rad1/Rad10-dependent processing of recombination intermediates.Mol Cell. 2008 May 9;30(3):325-35. doi: 10.1016/j.molcel.2008.02.028. Mol Cell. 2008. PMID: 18471978 Free PMC article.
-
Role of reciprocal exchange, one-ended invasion crossover and single-strand annealing on inverted and direct repeat recombination in yeast: different requirements for the RAD1, RAD10, and RAD52 genes.Genetics. 1995 Jan;139(1):109-23. doi: 10.1093/genetics/139.1.109. Genetics. 1995. PMID: 7705617 Free PMC article.
-
RAD1 and RAD10, but not other excision repair genes, are required for double-strand break-induced recombination in Saccharomyces cerevisiae.Mol Cell Biol. 1995 Apr;15(4):2245-51. doi: 10.1128/MCB.15.4.2245. Mol Cell Biol. 1995. PMID: 7891718 Free PMC article.
-
A tale of tails: insights into the coordination of 3' end processing during homologous recombination.Bioessays. 2009 Mar;31(3):315-21. doi: 10.1002/bies.200800195. Bioessays. 2009. PMID: 19260026 Free PMC article. Review.
-
Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair.Microbiol Mol Biol Rev. 2002 Dec;66(4):630-70, table of contents. doi: 10.1128/MMBR.66.4.630-670.2002. Microbiol Mol Biol Rev. 2002. PMID: 12456786 Free PMC article. Review.
Cited by
-
Inverted terminal repeats of adeno-associated virus decrease random integration of a gene targeting fragment in Saccharomyces cerevisiae.BMC Mol Biol. 2014 Feb 13;15:5. doi: 10.1186/1471-2199-15-5. BMC Mol Biol. 2014. PMID: 24521444 Free PMC article.
References
-
- Prado F, Cortes-Ledesma F, Huertas P, Aguilera A. Mitotic recombination in Saccharomyces cerevisiae. Curr Genet. 2003;42:185–198. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

