Solving a Hamiltonian Path Problem with a bacterial computer
- PMID: 19630940
- PMCID: PMC2723075
- DOI: 10.1186/1754-1611-3-11
Solving a Hamiltonian Path Problem with a bacterial computer
Abstract
Background: The Hamiltonian Path Problem asks whether there is a route in a directed graph from a beginning node to an ending node, visiting each node exactly once. The Hamiltonian Path Problem is NP complete, achieving surprising computational complexity with modest increases in size. This challenge has inspired researchers to broaden the definition of a computer. DNA computers have been developed that solve NP complete problems. Bacterial computers can be programmed by constructing genetic circuits to execute an algorithm that is responsive to the environment and whose result can be observed. Each bacterium can examine a solution to a mathematical problem and billions of them can explore billions of possible solutions. Bacterial computers can be automated, made responsive to selection, and reproduce themselves so that more processing capacity is applied to problems over time.
Results: We programmed bacteria with a genetic circuit that enables them to evaluate all possible paths in a directed graph in order to find a Hamiltonian path. We encoded a three node directed graph as DNA segments that were autonomously shuffled randomly inside bacteria by a Hin/hixC recombination system we previously adapted from Salmonella typhimurium for use in Escherichia coli. We represented nodes in the graph as linked halves of two different genes encoding red or green fluorescent proteins. Bacterial populations displayed phenotypes that reflected random ordering of edges in the graph. Individual bacterial clones that found a Hamiltonian path reported their success by fluorescing both red and green, resulting in yellow colonies. We used DNA sequencing to verify that the yellow phenotype resulted from genotypes that represented Hamiltonian path solutions, demonstrating that our bacterial computer functioned as expected.
Conclusion: We successfully designed, constructed, and tested a bacterial computer capable of finding a Hamiltonian path in a three node directed graph. This proof-of-concept experiment demonstrates that bacterial computing is a new way to address NP-complete problems using the inherent advantages of genetic systems. The results of our experiments also validate synthetic biology as a valuable approach to biological engineering. We designed and constructed basic parts, devices, and systems using synthetic biology principles of standardization and abstraction.
Figures

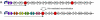
 and the 3' half is denoted by
and the 3' half is denoted by  . DNA edges are depicted by gene halves connected by arrows and flanked by triangles that represent hixC sites. Transcription in the direction of the solid arrow would terminate early and result in the expression of only one marker gene. b. Hin-mediated recombination would randomly reshuffle the DNA edges into many configurations. One possible example of an HPP solution configuration with its marker gene halves reunited is illustrated. Transcription in the direction of the solid arrow would result in expression of the six marker gene phenotypes.
. DNA edges are depicted by gene halves connected by arrows and flanked by triangles that represent hixC sites. Transcription in the direction of the solid arrow would terminate early and result in the expression of only one marker gene. b. Hin-mediated recombination would randomly reshuffle the DNA edges into many configurations. One possible example of an HPP solution configuration with its marker gene halves reunited is illustrated. Transcription in the direction of the solid arrow would result in expression of the six marker gene phenotypes.




References
-
- Karp RM. Reducibility among combinatorial problems. In: Miller RE, Thatcher JW, editor. Complexity of Computer Computations. Plenum Press; 1972. pp. 85–103.
-
- The international SAT competitions web page http://www.satcompetition.org/
-
- Gates W. Bounds for sorting by prefix reversal. Discrete Mathematics. 1979;27:47–57. doi: 10.1016/0012-365X(79)90068-2. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

