Distinct retroelement classes define evolutionary breakpoints demarcating sites of evolutionary novelty
- PMID: 19630942
- PMCID: PMC2736999
- DOI: 10.1186/1471-2164-10-334
Distinct retroelement classes define evolutionary breakpoints demarcating sites of evolutionary novelty
Abstract
Background: Large-scale genome rearrangements brought about by chromosome breaks underlie numerous inherited diseases, initiate or promote many cancers and are also associated with karyotype diversification during species evolution. Recent research has shown that these breakpoints are nonrandomly distributed throughout the mammalian genome and many, termed "evolutionary breakpoints" (EB), are specific genomic locations that are "reused" during karyotypic evolution. When the phylogenetic trajectory of orthologous chromosome segments is considered, many of these EB are coincident with ancient centromere activity as well as new centromere formation. While EB have been characterized as repeat-rich regions, it has not been determined whether specific sequences have been retained during evolution that would indicate previous centromere activity or a propensity for new centromere formation. Likewise, the conservation of specific sequence motifs or classes at EBs among divergent mammalian taxa has not been determined.
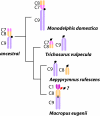
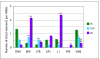
Results: To define conserved sequence features of EBs associated with centromere evolution, we performed comparative sequence analysis of more than 4.8 Mb within the tammar wallaby, Macropus eugenii, derived from centromeric regions (CEN), euchromatic regions (EU), and an evolutionary breakpoint (EB) that has undergone convergent breakpoint reuse and past centromere activity in marsupials. We found a dramatic enrichment for long interspersed nucleotide elements (LINE1s) and endogenous retroviruses (ERVs) and a depletion of short interspersed nucleotide elements (SINEs) shared between CEN and EBs. We analyzed the orthologous human EB (14q32.33), known to be associated with translocations in many cancers including multiple myelomas and plasma cell leukemias, and found a conserved distribution of similar repetitive elements.
Conclusion: Our data indicate that EBs tracked within the class Mammalia harbor sequence features retained since the divergence of marsupials and eutherians that may have predisposed these genomic regions to large-scale chromosomal instability.
Figures







References
-
- Padilla-Nash H, Heselmeyer-Haddad K, Wangsa D, Zhang H, Ghadimi B, Macville M, Augustus M, Schröck E, Hilgenfeld E, Ried T. Jumping translocations are common in solid tumor cell lines and result in recurrent fusions of whole chromosome arms. Genes Chromosomes Cancer. 2001;30:349–363. doi: 10.1002/gcc.1101. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

