RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway
- PMID: 19631370
- PMCID: PMC2747301
- DOI: 10.1016/j.cell.2009.06.015
RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway
Abstract
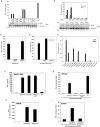
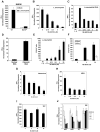
Type I interferons (IFNs) are important for antiviral and autoimmune responses. Retinoic acid-induced gene I (RIG-I) and mitochondrial antiviral signaling (MAVS) proteins mediate IFN production in response to cytosolic double-stranded RNA or single-stranded RNA containing 5'-triphosphate (5'-ppp). Cytosolic B form double-stranded DNA, such as poly(dA-dT)*poly(dA-dT) [poly(dA-dT)], can also induce IFN-beta, but the underlying mechanism is unknown. Here, we show that the cytosolic poly(dA-dT) DNA is converted into 5'-ppp RNA to induce IFN-beta through the RIG-I pathway. Biochemical purification led to the identification of DNA-dependent RNA polymerase III (Pol-III) as the enzyme responsible for synthesizing 5'-ppp RNA from the poly(dA-dT) template. Inhibition of RNA Pol-III prevents IFN-beta induction by transfection of DNA or infection with DNA viruses. Furthermore, Pol-III inhibition abrogates IFN-beta induction by the intracellular bacterium Legionella pneumophila and promotes the bacterial growth. These results suggest that RNA Pol-III is a cytosolic DNA sensor involved in innate immune responses.
Figures





Comment in
-
DNA makes RNA makes innate immunity.Cell. 2009 Aug 7;138(3):428-30. doi: 10.1016/j.cell.2009.07.021. Cell. 2009. PMID: 19665965
References
-
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. - PubMed
-
- Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. - PubMed
-
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

