Structure and function of Na(+)-symporters with inverted repeats
- PMID: 19631523
- PMCID: PMC3496787
- DOI: 10.1016/j.sbi.2009.06.002
Structure and function of Na(+)-symporters with inverted repeats
Abstract
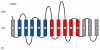
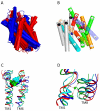
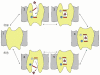
Symporters are membrane proteins that couple energy stored in electrochemical potential gradients to drive the cotransport of molecules and ions into cells. Traditionally, proteins are classified into gene families based on sequence homology and functional properties, for example the sodium glucose (SLC5 or Sodium Solute Symporter Family, SSS or SSF) and GABA (SLC6 or Neurotransmitter Sodium Symporter Family, NSS or SNF) symporter families [1-4]. Recently, it has been established that four Na(+)-symporter proteins with unrelated sequences have a common structural core containing an inverted repeat of 5 transmembrane (TM) helices [5(**)-8(**)]. Analysis of these four structures reveals that they reside in different conformations along the transport cycle providing atomic insight into the mechanism of sodium solute cotransport.
Figures

References
-
- Nelson N. The Family of Na+/C1- Neuortransmitter Transporters. Journal of Neurochemistry. 199871:1786–1803. - PubMed
-
- Wright EMaT E. The Sodium Glucose Cotransport Family SLC5. Pflugers Arch. 2004:510–518. - PubMed
-
- Turk E, Wright EM. Membrane topology motifs in the SGLT cotransporter family. J Membr Biol. 1997;159:1–20. - PubMed
-
- Chen NH, Reith ME, Quick MW. Synaptic uptake and beyond: the sodium-and chloride-dependent neurotransmitter transporter family SLC6. Pflugers Arch. 2004;447:519–531. - PubMed
-
- Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl--dependent neurotransmitter transporters. Nature. 2005;437:215–223. - PubMed
-
The first high resolution crystal structure of a sodium symporter with the inverted 5TM motif containing a pair of transmembrane discontinuous helices. The substrate, leucine, and one sodium was bound in a occluded site halfway across the membrane close to these two discontinuous helices. A route for substrate entry to the binding site from the external solution was identified and this conformation of the protein was termed the outward facing substrate occluded conformation.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

