DNA polymerase family X: function, structure, and cellular roles
- PMID: 19631767
- PMCID: PMC2846199
- DOI: 10.1016/j.bbapap.2009.07.008
DNA polymerase family X: function, structure, and cellular roles
Abstract
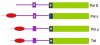
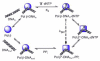
The X family of DNA polymerases in eukaryotic cells consists of terminal transferase and DNA polymerases beta, lambda, and mu. These enzymes have similar structural portraits, yet different biochemical properties, especially in their interactions with DNA. None of these enzymes possesses a proofreading subdomain, and their intrinsic fidelity of DNA synthesis is much lower than that of a polymerase that functions in cellular DNA replication. In this review, we discuss the similarities and differences of three members of Family X: polymerases beta, lambda, and mu. We focus on biochemical mechanisms, structural variation, fidelity and lesion bypass mechanisms, and cellular roles. Remarkably, although these enzymes have similar three-dimensional structures, their biochemical properties and cellular functions differ in important ways that impact cellular function.
Copyright (c) 2010 Elsevier B.V. All rights reserved.
Figures
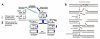

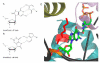

References
-
- Chang LM, Bollum FJ. Low molecular weight deoxyribonucleic acid polymerase in mammalian cells. J Biol Chem. 1971;246:5835–5837. - PubMed
-
- Li F, Xiong Y, Wang J, Cho HD, Tomita K, Weiner AM, Steitz TA. Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and its complexes with ATP or CTP. Cell. 2002;111:815–824. - PubMed
-
- Uchiyama Y, Takeuchi R, Kodera H, Sakaguchi K. Distribution and roles of X-family DNA polymerases in eukaryotes. Biochimie. 2009;91:165–170. - PubMed
-
- Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–476. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

