Peptides in the treatment of AIDS
- PMID: 19632107
- PMCID: PMC2763535
- DOI: 10.1016/j.sbi.2009.07.003
Peptides in the treatment of AIDS
Abstract
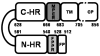
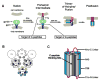
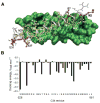
Fusion of HIV-1 and target cells is mediated by the envelope protein gp41 that undergoes a series of conformational changes during the process of infection. Knowledge of the structural biology of gp41 allows the design of potent peptide inhibitors that prevent the virus from entering lymphocytes and macrophages. The design of such inhibitors is the subject of this review.
Figures

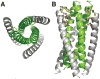
Similar articles
-
Development of HIV-1 fusion inhibitors targeting gp41.Curr Med Chem. 2014 Jun;21(17):1976-96. doi: 10.2174/0929867321666131218094559. Curr Med Chem. 2014. PMID: 24350848 Review.
-
Peptide and non-peptide HIV fusion inhibitors.Curr Pharm Des. 2002;8(8):563-80. doi: 10.2174/1381612024607180. Curr Pharm Des. 2002. PMID: 11945159 Review.
-
Novel therapies based on mechanisms of HIV-1 cell entry.N Engl J Med. 2003 May 29;348(22):2228-38. doi: 10.1056/NEJMra022812. N Engl J Med. 2003. PMID: 12773651 Review. No abstract available.
-
Development of small molecule HIV-1 fusion inhibitors: linking biology to chemistry.Curr Pharm Des. 2013;19(10):1827-34. doi: 10.2174/1381612811319100007. Curr Pharm Des. 2013. PMID: 23092276 Review.
-
Qadirvirtide.Pak J Pharm Sci. 2011 Oct;24(4):593-5. Pak J Pharm Sci. 2011. PMID: 21959827 Review.
Cited by
-
Biophysical property and broad anti-HIV activity of albuvirtide, a 3-maleimimidopropionic acid-modified peptide fusion inhibitor.PLoS One. 2012;7(3):e32599. doi: 10.1371/journal.pone.0032599. Epub 2012 Mar 5. PLoS One. 2012. PMID: 22403678 Free PMC article.
-
Prediction of the interaction of HIV-1 integrase and its dicaffeoylquinic acid inhibitor through molecular modeling approach.Ethn Dis. 2010 Winter;20(1 Suppl 1):S1-45-9. Ethn Dis. 2010. PMID: 20521384 Free PMC article.
-
Design, synthesis, and evaluation of indole compounds as novel inhibitors targeting Gp41.Bioorg Med Chem Lett. 2010 Mar 1;20(5):1500-3. doi: 10.1016/j.bmcl.2010.01.111. Epub 2010 Jan 25. Bioorg Med Chem Lett. 2010. PMID: 20153190 Free PMC article.
-
A targeted covalent small molecule inhibitor of HIV-1 fusion.Chem Commun (Camb). 2021 May 6;57(37):4528-4531. doi: 10.1039/d1cc01013a. Chem Commun (Camb). 2021. PMID: 33956029 Free PMC article.
-
Anti-antimicrobial peptides: folding-mediated host defense antagonists.J Biol Chem. 2013 Jul 12;288(28):20162-72. doi: 10.1074/jbc.M113.459560. Epub 2013 Jun 4. J Biol Chem. 2013. PMID: 23737519 Free PMC article.
References
-
- Liu S, Wu S, Jiang S. HIV entry inhibitors targeting gp41: from polypeptides to small-molecule compounds. Curr Pharm Des. 2007;13:143–162. Comprehensive review of the design of HIV-1 inhibitors targeting gp41 (133 references) - PubMed
-
- Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. Determination of the crystal structure of the gp41 core formed using synthetic peptides. - PubMed
-
- Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. Description of the crystal structure of the gp41 core using crystals formed from biosynthetic and synthetic peptides. - PubMed
-
- Chan DC, Kim PS. HIV entry and its inhibition. Cell. 1998;93:681–684. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

