Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer
- PMID: 19632176
- PMCID: PMC2726827
- DOI: 10.1016/j.cell.2009.04.056
Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer
Abstract
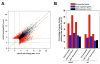
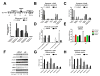
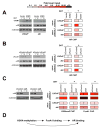
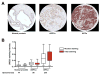
The evolution of prostate cancer from an androgen-dependent state to one that is androgen-independent marks its lethal progression. The androgen receptor (AR) is essential in both, though its function in androgen-independent cancers is poorly understood. We have defined the direct AR-dependent target genes in both androgen-dependent and -independent cancer cells by generating AR-dependent gene expression profiles and AR cistromes. In contrast to what is found in androgen-dependent cells, AR selectively upregulates M-phase cell-cycle genes in androgen-independent cells, including UBE2C, a gene that inactivates the M-phase checkpoint. We find that epigenetic marks at the UBE2C enhancer, notably histone H3K4 methylation and FoxA1 transcription factor binding, are present in androgen-independent cells and direct AR-enhancer binding and UBE2C activation. Thus, the role of AR in androgen-independent cancer cells is not to direct the androgen-dependent gene expression program without androgen, but rather to execute a distinct program resulting in androgen-independent growth.
Figures



References
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell . 2005;120:169–181. - PubMed
-
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. - PubMed
-
- Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, Rosenfeld MG, Sawyers CL. Molecular determinants of resistance to antiandrogen therapy. Nat Med. 2004;10:33–39. - PubMed
-
- Comstock CE, Knudsen KE. The complex role of AR signaling after cytotoxic insult: implications for cell-cycle-based chemotherapeutics. Cell Cycle. 2007;6:1307–1313. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

