Comparative sequence analysis of Mycobacterium leprae and the new leprosy-causing Mycobacterium lepromatosis
- PMID: 19633074
- PMCID: PMC2747882
- DOI: 10.1128/JB.00762-09
Comparative sequence analysis of Mycobacterium leprae and the new leprosy-causing Mycobacterium lepromatosis
Abstract
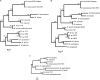
Mycobacterium lepromatosis is a newly discovered leprosy-causing organism. Preliminary phylogenetic analysis of its 16S rRNA gene and a few other gene segments revealed significant divergence from Mycobacterium leprae, a well-known cause of leprosy, that justifies the status of M. lepromatosis as a new species. In this study we analyzed the sequences of 20 genes and pseudogenes (22,814 nucleotides). Overall, the level of matching of these sequences with M. leprae sequences was 90.9%, which substantiated the species-level difference; the levels of matching for the 16S rRNA genes and 14 protein-encoding genes were 98.0% and 93.1%, respectively, but the level of matching for five pseudogenes was only 79.1%. Five conserved protein-encoding genes were selected to construct phylogenetic trees and to calculate the numbers of synonymous substitutions (dS values) and nonsynonymous substitutions (dN values) in the two species. Robust phylogenetic trees constructed using concatenated alignment of these genes placed M. lepromatosis and M. leprae in a tight cluster with long terminal branches, implying that the divergence occurred long ago. The dS and dN values were also much higher than those for other closest pairs of mycobacteria. The dS values were 14 to 28% of the dS values for M. leprae and Mycobacterium tuberculosis, a more divergent pair of species. These results thus indicate that M. lepromatosis and M. leprae diverged approximately 10 million years ago. The M. lepromatosis pseudogenes analyzed that were also pseudogenes in M. leprae showed nearly neutral evolution, and their relative ages were similar to those of M. leprae pseudogenes, suggesting that they were pseudogenes before divergence. Taken together, the results described above indicate that M. lepromatosis and M. leprae diverged from a common ancestor after the massive gene inactivation event described previously for M. leprae.
Figures
References
-
- Adekambi, T., T. M. Shinnick, D. Raoult, and M. Drancourt. 2008. Complete rpoB gene sequencing as a suitable supplement to DNA-DNA hybridization for bacterial species and genus delineation. Int. J. Syst. Evol. Microbiol. 58:1807-1814. - PubMed
-
- Canback, B., I. Tamas, and S. G. Andersson. 2004. A phylogenomic study of endosymbiotic bacteria. Mol. Biol. Evol. 21:1110-1122. - PubMed
-
- Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. - PubMed
-
- Cole, S. T., K. Eiglmeier, J. Parkhill, K. D. James, N. R. Thomson, P. R. Wheeler, N. Honore, T. Garnier, C. Churcher, D. Harris, K. Mungall, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. M. Davies, K. Devlin, S. Duthoy, T. Feltwell, A. Fraser, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, C. Lacroix, J. Maclean, S. Moule, L. Murphy, K. Oliver, M. A. Quail, M. A. Rajandream, K. M. Rutherford, S. Rutter, K. Seeger, S. Simon, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, K. Taylor, S. Whitehead, J. R. Woodward, and B. G. Barrell. 2001. Massive gene decay in the leprosy bacillus. Nature 409:1007-1011. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

