Characterization of novel alleles of the Escherichia coli umuDC genes identifies additional interaction sites of UmuC with the beta clamp
- PMID: 19633075
- PMCID: PMC2747890
- DOI: 10.1128/JB.00292-09
Characterization of novel alleles of the Escherichia coli umuDC genes identifies additional interaction sites of UmuC with the beta clamp
Abstract
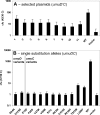
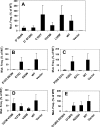
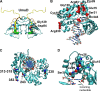
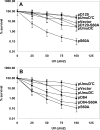
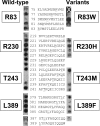
Translesion synthesis is a DNA damage tolerance mechanism by which damaged DNA in a cell can be replicated by specialized DNA polymerases without being repaired. The Escherichia coli umuDC gene products, UmuC and the cleaved form of UmuD, UmuD', comprise a specialized, potentially mutagenic translesion DNA polymerase, polymerase V (UmuD'(2)C). The full-length UmuD protein, together with UmuC, plays a role in a primitive DNA damage checkpoint by decreasing the rate of DNA synthesis. It has been proposed that the checkpoint is manifested as a cold-sensitive phenotype that is observed when the umuDC gene products are overexpressed. Elevated levels of the beta processivity clamp along with elevated levels of the umuDC gene products, UmuD'C, exacerbate the cold-sensitive phenotype. We used this observation as the basis for genetic selection to identify two alleles of umuD' and seven alleles of umuC that do not exacerbate the cold-sensitive phenotype when they are present in cells with elevated levels of the beta clamp. The variants were characterized to determine their abilities to confer the umuD'C-specific phenotype UV-induced mutagenesis. The umuD variants were assayed to determine their proficiencies in UmuD cleavage, and one variant (G129S) rendered UmuD noncleaveable. We found at least two UmuC residues, T243 and L389, that may further define the beta binding region on UmuC. We also identified UmuC S31, which is predicted to bind to the template nucleotide, as a residue that is important for UV-induced mutagenesis.
Figures
References
-
- Becherel, O. J., R. P. P. Fuchs, and J. Wagner. 2002. Pivotal role of the β-clamp in translesion DNA synthesis and mutagenesis in E. coli cells. DNA Repair 1:703-708. - PubMed
-
- Beuning, P. J., D. Sawicka, D. Barsky, and G. C. Walker. 2006. Two processivity clamp interactions differentially alter the dual activities of UmuC. Mol. Microbiol. 59:460-474. - PubMed
-
- Beuning, P. J., S. M. Simon, V. G. Godoy, D. F. Jarosz, and G. C. Walker. 2006. Characterization of Escherichia coli translesion synthesis polymerases and their accessory factors. Methods Enzymol. 408:318-340. - PubMed
-
- Beuning, P. J., S. M. Simon, A. Zemla, D. Barsky, and G. C. Walker. 2006. A non-cleavable UmuD variant that acts as a UmuD′ mimic. J. Biol. Chem. 281:9633-9640. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

