Potential role of group IIC-attC introns in integron cassette formation
- PMID: 19633079
- PMCID: PMC2747902
- DOI: 10.1128/JB.00674-09
Potential role of group IIC-attC introns in integron cassette formation
Abstract
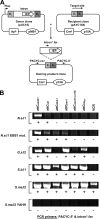
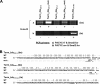
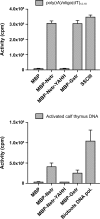
Integrons are natural expression vectors in which gene cassettes are integrated downstream of a promoter region by a site-specific recombinase. Gene cassettes usually consist of a single gene followed by a recombination site designated attC. A major unanswered question is how a gene becomes associated with an attC site. Here, we investigate the potential role of a specific lineage of group IIC introns, named group IIC-attC, in cassette formation. Group IIC-attC introns preferentially target attC while retaining the ability to target transcriptional terminators. We show using a PCR-based mobility assay with Escherichia coli that the S.ma.I2 intron from the genome of a clinical isolate of Serratia marcescens can target both attC site and putative terminator motifs of resistance genes. Quantitative results showed that S.ma.I2 is more efficient in targeting various attC sequences than three group IIC-attC introns (54 to 64% sequence identity) from the genomes of environmental isolates. We also show that purified group IIC-attC intron-encoded reverse transcriptases have both RNA-dependent and DNA-dependent DNA polymerase activities in vitro. These data permit us to suggest a new model for gene cassette formation, in which a group IIC-attC intron targets separately a transcriptional terminator adjoining a gene and an isolated attC, joins the gene and the attC by homologous recombination, and then splices and reverse transcribes a gene-attC RNA template, leading to the formation of a cassette.
Figures


References
-
- Boucher, Y., M. Labbate, J. E. Koenig, and H. W. Stokes. 2007. Integrons: mobilizable platforms that promote genetic diversity in bacteria. Trends Microbiol. 15:301-309. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

