Characterization and comparative analysis of the genes encoding Haemophilus parasuis outer membrane proteins P2 and P5
- PMID: 19633080
- PMCID: PMC2747881
- DOI: 10.1128/JB.00469-09
Characterization and comparative analysis of the genes encoding Haemophilus parasuis outer membrane proteins P2 and P5
Abstract
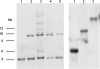
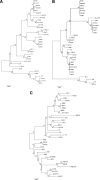
Haemophilus parasuis is a swine pathogen of significant industry concern, but little is known about how the organism causes disease. A related human pathogen, Haemophilus influenzae, has been better studied, and many of its virulence factors have been identified. Two of these, outer membrane proteins P2 and P5, are known to have important virulence properties. The goals of this study were to identify, analyze, and compare the genetic relatedness of orthologous genes encoding P2 and P5 proteins in a diverse group of 35 H. parasuis strains. Genes encoding P2 and P5 proteins were detected in all H. parasuis strains evaluated. The predicted amino acid sequences for both P2 and P5 proteins exhibit considerable heterogeneity, particularly in regions corresponding to predicted extracellular loops. Twenty-five variants of P2 and 17 variants of P5 were identified. The P2 proteins of seven strains were predicted to contain a highly conserved additional extracellular loop compared to the remaining strains and to H. influenzae P2. Antigenic-site predictions coincided with predicted extracellular loop regions of both P2 and P5. Neighbor-joining trees constructed using P2 and P5 sequences predicted divergent evolutionary histories distinct from those predicted by a multilocus sequence typing phylogeny based on partial sequencing of seven housekeeping genes. Real-time reverse transcription-PCR indicated that both genes are expressed in all of the strains.
Figures







References
-
- Andersen, C., E. Maier, G. Kemmer, J. Blass, A. K. Hilpert, R. Benz, and J. Reidl. 2003. Porin OmpP2 of Haemophilus influenzae shows specificity for nicotinamide-derived nucleotide substrates. J. Biol. Chem. 278:24269-24276. - PubMed
-
- Bigas, A., M. E. Garrido, A. M. de Rozas, I. Badiola, J. Barbe, and M. Llagostera. 2005. Development of a genetic manipulation system for Haemophilus parasuis. Vet. Microbiol. 105:223-228. - PubMed
-
- Bouchet, B., G. Vanier, M. Jacques, E. Auger, and M. Gottschalk. 2009. Studies on the interactions of Haemophilus parasuis with porcine epithelial tracheal cells: limited role of LOS in apoptosis and pro-inflammatory cytokine release. Microb. Pathog. 46:108-113. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

