Feedback control of DnaA-mediated replication initiation by replisome-associated HdaA protein in Caulobacter
- PMID: 19633089
- PMCID: PMC2737981
- DOI: 10.1128/JB.00525-09
Feedback control of DnaA-mediated replication initiation by replisome-associated HdaA protein in Caulobacter
Abstract
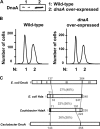
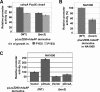
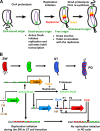
Chromosome replication in Caulobacter crescentus is tightly regulated to ensure that initiation occurs at the right time and only once during the cell cycle. The timing of replication initiation is controlled by both CtrA and DnaA. CtrA binds to and silences the origin. Upon the clearance of CtrA from the cell, the DnaA protein accumulates and allows loading of the replisome at the origin. Here, we identify an additional layer of replication initiation control that is mediated by the HdaA protein. In Escherichia coli, the Hda protein inactivates DnaA after replication initiation. We show that the Caulobacter HdaA homologue is necessary to restrict the initiation of DNA replication to only once per cell cycle and that it dynamically colocalizes with the replisome throughout the cell cycle. Moreover, the transcription of hdaA is directly activated by DnaA, providing a robust feedback regulatory mechanism that adjusts the levels of HdaA to inactivate DnaA.
Figures



References
-
- Bastedo, D. P., and G. T. Marczynski. 2009. CtrA response regulator binding to the Caulobacter chromosome replication origin is required during nutrient and antibiotic stress as well as during cell cycle progression. Mol. Microbiol. 72139-154. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

