Genome-wide identification of Saccharomyces cerevisiae genes required for maximal tolerance to ethanol
- PMID: 19633105
- PMCID: PMC2747848
- DOI: 10.1128/AEM.00845-09
Genome-wide identification of Saccharomyces cerevisiae genes required for maximal tolerance to ethanol
Abstract
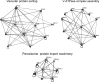
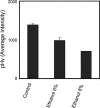
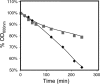
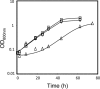
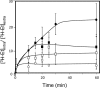
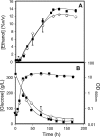
The understanding of the molecular basis of yeast resistance to ethanol may guide the design of rational strategies to increase process performance in industrial alcoholic fermentations. In this study, the yeast disruptome was screened for mutants with differential susceptibility to stress induced by high ethanol concentrations in minimal growth medium. Over 250 determinants of resistance to ethanol were identified. The most significant gene ontology terms enriched in this data set are those associated with intracellular organization, biogenesis, and transport, in particular, regarding the vacuole, the peroxisome, the endosome, and the cytoskeleton, and those associated with the transcriptional machinery. Clustering the proteins encoded by the identified determinants of ethanol resistance by their known physical and genetic interactions highlighted the importance of the vacuolar protein sorting machinery, the vacuolar H(+)-ATPase complex, and the peroxisome protein import machinery. Evidence showing that vacuolar acidification and increased resistance to the cell wall lytic enzyme beta-glucanase occur in response to ethanol-induced stress was obtained. Based on the genome-wide results, the particular role of the FPS1 gene, encoding a plasma membrane aquaglyceroporin which mediates controlled glycerol efflux, in ethanol stress resistance was further investigated. FPS1 expression contributes to decreased [(3)H]ethanol accumulation in yeast cells, suggesting that Fps1p may also play a role in maintaining the intracellular ethanol level during active fermentation. The increased expression of FPS1 confirmed the important role of this gene in alcoholic fermentation, leading to increased final ethanol concentration under conditions that lead to high ethanol production.
Figures

Similar articles
-
Identification of genes required for maximal tolerance to high-glucose concentrations, as those present in industrial alcoholic fermentation media, through a chemogenomics approach.OMICS. 2010 Apr;14(2):201-10. doi: 10.1089/omi.2009.0149. OMICS. 2010. PMID: 20210661
-
Vacuolar H+-ATPase Protects Saccharomyces cerevisiae Cells against Ethanol-Induced Oxidative and Cell Wall Stresses.Appl Environ Microbiol. 2016 May 2;82(10):3121-3130. doi: 10.1128/AEM.00376-16. Print 2016 May 15. Appl Environ Microbiol. 2016. PMID: 26994074 Free PMC article.
-
Increased expression of the yeast multidrug resistance ABC transporter Pdr18 leads to increased ethanol tolerance and ethanol production in high gravity alcoholic fermentation.Microb Cell Fact. 2012 Jul 27;11:98. doi: 10.1186/1475-2859-11-98. Microb Cell Fact. 2012. PMID: 22839110 Free PMC article.
-
The ethanol stress response and ethanol tolerance of Saccharomyces cerevisiae.J Appl Microbiol. 2010 Jul;109(1):13-24. doi: 10.1111/j.1365-2672.2009.04657.x. Epub 2010 Jan 11. J Appl Microbiol. 2010. PMID: 20070446 Review.
-
Omics analysis of acetic acid tolerance in Saccharomyces cerevisiae.World J Microbiol Biotechnol. 2017 May;33(5):94. doi: 10.1007/s11274-017-2259-9. Epub 2017 Apr 12. World J Microbiol Biotechnol. 2017. PMID: 28405910 Review.
Cited by
-
Traditional Norwegian Kveik Are a Genetically Distinct Group of Domesticated Saccharomyces cerevisiae Brewing Yeasts.Front Microbiol. 2018 Sep 12;9:2137. doi: 10.3389/fmicb.2018.02137. eCollection 2018. Front Microbiol. 2018. PMID: 30258422 Free PMC article.
-
Current Ethanol Production Requirements for the Yeast Saccharomyces cerevisiae.Int J Microbiol. 2022 Aug 13;2022:7878830. doi: 10.1155/2022/7878830. eCollection 2022. Int J Microbiol. 2022. PMID: 35996633 Free PMC article. Review.
-
Condition-specific promoter activities in Saccharomyces cerevisiae.Microb Cell Fact. 2018 Apr 10;17(1):58. doi: 10.1186/s12934-018-0899-6. Microb Cell Fact. 2018. PMID: 29631591 Free PMC article.
-
Genetic improvement of native xylose-fermenting yeasts for ethanol production.J Ind Microbiol Biotechnol. 2015 Jan;42(1):1-20. doi: 10.1007/s10295-014-1535-z. Epub 2014 Nov 18. J Ind Microbiol Biotechnol. 2015. PMID: 25404205 Review.
-
Adaptation to High Ethanol Reveals Complex Evolutionary Pathways.PLoS Genet. 2015 Nov 6;11(11):e1005635. doi: 10.1371/journal.pgen.1005635. eCollection 2015 Nov. PLoS Genet. 2015. PMID: 26545090 Free PMC article.
References
-
- Aguilera, F., R. A. Peinado, C. Millan, J. M. Ortega, and J. C. Mauricio. 2006. Relationship between ethanol tolerance, H+-ATPase activity and the lipid composition of the plasma membrane in different wine yeast strains. Int. J. Food Microbiol. 110:34-42. - PubMed
-
- Alexandre, H., V. Ansanay-Galeote, S. Dequin, and B. Blondin. 2001. Global gene expression during short-term ethanol stress in Saccharomyces cerevisiae. FEBS Lett. 498:98-103. - PubMed
-
- Alper, H., J. Moxley, E. Nevoigt, G. R. Fink, and G. Stephanopoulos. 2006. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science 314:1565-1568. - PubMed
-
- Carmelo, V., H. Santos, and I. Sá-Correia. 1997. Effect of extracellular acidification on the activity of plasma membrane ATPase and on the cytosolic and vacuolar pH of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1325:63-70. - PubMed
-
- Casey, G. P., and W. M. Ingledew. 1986. Ethanol tolerance in yeasts. Crit. Rev. Microbiol. 13:219-280. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

