Increased malonyl coenzyme A biosynthesis by tuning the Escherichia coli metabolic network and its application to flavanone production
- PMID: 19633125
- PMCID: PMC2747866
- DOI: 10.1128/AEM.00270-09
Increased malonyl coenzyme A biosynthesis by tuning the Escherichia coli metabolic network and its application to flavanone production
Abstract
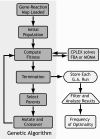
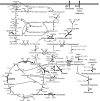
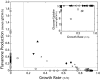
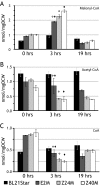
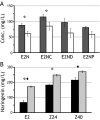
Identification of genetic targets able to bring about changes to the metabolite profiles of microorganisms continues to be a challenging task. We have independently developed a cipher of evolutionary design (CiED) to identify genetic perturbations, such as gene deletions and other network modifications, that result in optimal phenotypes for the production of end products, such as recombinant natural products. Coupled to an evolutionary search, our method demonstrates the utility of a purely stoichiometric network to predict improved Escherichia coli genotypes that more effectively channel carbon flux toward malonyl coenzyme A (CoA) and other cofactors in an effort to generate recombinant strains with enhanced flavonoid production capacity. The engineered E. coli strains were constructed first by the targeted deletion of native genes predicted by CiED and then second by incorporating selected overexpressions, including those of genes required for the coexpression of the plant-derived flavanones, acetate assimilation, acetyl-CoA carboxylase, and the biosynthesis of coenzyme A. As a result, the specific flavanone production from our optimally engineered strains was increased by over 660% for naringenin (15 to 100 mg/liter/optical density unit [OD]) and by over 420% for eriodictyol (13 to 55 mg/liter/OD).
Figures
References
-
- Alper, H., Y. S. Jin, J. F. Moxley, and G. Stephanopoulos. 2005. Identifying gene targets for the metabolic engineering of lycopene biosynthesis in Escherichia coli. Metab. Eng. 7:155-164. - PubMed
-
- Alper, H., K. Miyaoku, and G. Stephanopoulos. 2005. Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets. Nat. Biotechnol. 23:612-616. - PubMed
-
- Atsumi, S., A. F. Cann, M. Connor, C. R. Shen, K. M. Smith, M. P. Brynildsen, K. J. Y. Chou, T. Hanai, and J. C. Liao. 2008. Metabolic engineering of Escherichia coli for 1-butanol production. Metab. Eng. 10:305-311. - PubMed
-
- Barkovich, R., and J. C. Liao. 2001. Metabolic engineering of isoprenoids. Metab. Eng. 3:27-39. - PubMed
-
- Burgard, A. P., P. Pharkya, and C. D. Maranas. 2003. Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol. Bioeng. 84:647-657. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

