Targeted disruption of Zfp36l2, encoding a CCCH tandem zinc finger RNA-binding protein, results in defective hematopoiesis
- PMID: 19633199
- PMCID: PMC2746470
- DOI: 10.1182/blood-2009-04-214619
Targeted disruption of Zfp36l2, encoding a CCCH tandem zinc finger RNA-binding protein, results in defective hematopoiesis
Abstract
Members of the tristetraprolin family of tandem CCCH finger proteins can bind to AU-rich elements in the 3'-untranslated region of mRNAs, leading to their deadenylation and subsequent degradation. Partial deficiency of 1 of the 4 mouse tristetraprolin family members, Zfp36l2, resulted in complete female infertility because of early embryo death. We have now generated mice completely deficient in the ZFP36L2 protein. Homozygous Zfp36l2 knockout (KO) mice died within approximately 2 weeks of birth, apparently from intestinal or other hemorrhage. Analysis of peripheral blood from KO mice showed a decrease in red and white cells, hemoglobin, hematocrit, and platelets. Yolk sacs from embryonic day 11.5 (E11.5) Zfp36l2 KO mice and fetal livers from E14.5 KO mice gave rise to markedly reduced numbers of definitive multilineage and lineage-committed hematopoietic progenitors. Competitive reconstitution experiments demonstrated that Zfp36l2 KO fetal liver hematopoietic stem cells were unable to adequately reconstitute the hematopoietic system of lethally irradiated recipients. These data establish Zfp36l2 as a critical modulator of definitive hematopoiesis and suggest a novel regulatory pathway involving control of mRNA stability in the life cycle of hematopoietic stem and progenitor cells.
Figures

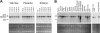
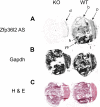
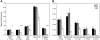

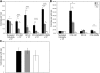
References
-
- Lai WS, Stumpo DJ, Blackshear PJ. Rapid insulin-stimulated accumulation of an mRNA encoding a proline-rich protein. J Biol Chem. 1990;265(27):16556–16563. - PubMed
-
- Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281(5379):1001–1005. - PubMed
-
- Taylor GA, Carballo E, Lee DM, et al. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4(5):445–454. - PubMed
-
- Carballo E, Blackshear PJ. Roles of tumor necrosis factor-alpha receptor subtypes in the pathogenesis of the tristetraprolin-deficiency syndrome. Blood. 2001;98(8):2389–2395. - PubMed
-
- Carballo E, Lai WS, Blackshear PJ. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood. 2000;95(6):1891–1899. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
