A genetic screen for nitrate regulatory mutants captures the nitrate transporter gene NRT1.1
- PMID: 19633234
- PMCID: PMC2735993
- DOI: 10.1104/pp.109.140434
A genetic screen for nitrate regulatory mutants captures the nitrate transporter gene NRT1.1
Abstract
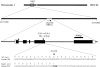
Nitrate regulatory mutants (nrg) of Arabidopsis (Arabidopsis thaliana) were sought using a genetic screen that employed a nitrate-inducible promoter fused to the yellow fluorescent protein marker gene YFP. A mutation was identified that impaired nitrate induction, and it was localized to the nitrate regulatory gene NLP7, demonstrating the validity of this screen. A second, independent mutation (nrg1) mapped to a region containing the NRT1.1 (CHL1) nitrate transporter gene on chromosome 1. Sequence analysis of NRT1.1 in the mutant revealed a nonsense mutation that truncated the NRT1.1 protein at amino acid 301. The nrg1 mutation disrupted nitrate regulation of several endogenous genes as induction of three nitrate-responsive genes (NIA1, NiR, and NRT2.1) was dramatically reduced in roots of the mutant after 2-h treatment using nitrate concentrations from 0.25 to 20 mm. Another nrt1.1 mutant (deletion mutant chl1-5) showed a similar phenotype. The loss of nitrate induction in the two nrt1.1 mutants (nrg1 and chl1-5) was not explained by reduced nitrate uptake and was reversed by nitrogen deprivation. Microarray analysis showed that nitrate induction of 111 genes was reduced and of three genes increased 2-fold or more in the nrg1 mutant. Genes involved in nitrate assimilation, energy metabolism, and pentose-phosphate pathway were most affected. These results strongly support the model that NRT1.1 acts as a nitrate regulator or sensor in Arabidopsis.
Figures

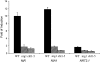

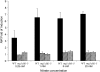

Similar articles
-
Transcript profiling in the chl1-5 mutant of Arabidopsis reveals a role of the nitrate transporter NRT1.1 in the regulation of another nitrate transporter, NRT2.1.Plant Cell. 2004 Sep;16(9):2433-47. doi: 10.1105/tpc.104.024380. Epub 2004 Aug 19. Plant Cell. 2004. PMID: 15319483 Free PMC article.
-
The Arabidopsis NLP7 gene regulates nitrate signaling via NRT1.1-dependent pathway in the presence of ammonium.Sci Rep. 2018 Jan 24;8(1):1487. doi: 10.1038/s41598-018-20038-4. Sci Rep. 2018. PMID: 29367694 Free PMC article.
-
A reevaluation of the contribution of NRT1.1 to nitrate uptake in Arabidopsis under low-nitrate supply.FEBS Lett. 2019 Aug;593(15):2051-2059. doi: 10.1002/1873-3468.13473. Epub 2019 Jun 13. FEBS Lett. 2019. PMID: 31172512
-
Nitrate transceptor(s) in plants.J Exp Bot. 2011 Apr;62(7):2299-308. doi: 10.1093/jxb/erq419. Epub 2011 Jan 14. J Exp Bot. 2011. PMID: 21239382 Review.
-
The primary nitrate response: a multifaceted signalling pathway.J Exp Bot. 2014 Oct;65(19):5567-76. doi: 10.1093/jxb/eru245. Epub 2014 Jun 18. J Exp Bot. 2014. PMID: 24942915 Review.
Cited by
-
HBI transcription factor-mediated ROS homeostasis regulates nitrate signal transduction.Plant Cell. 2021 Sep 24;33(9):3004-3021. doi: 10.1093/plcell/koab165. Plant Cell. 2021. PMID: 34129038 Free PMC article.
-
Multiple regulatory elements in the Arabidopsis NIA1 promoter act synergistically to form a nitrate enhancer.Plant Physiol. 2010 Sep;154(1):423-32. doi: 10.1104/pp.110.162586. Epub 2010 Jul 28. Plant Physiol. 2010. PMID: 20668061 Free PMC article.
-
An updated model for nitrate uptake modelling in plants. I. Functional component: cross-combination of flow-force interpretation of nitrate uptake isotherms, and environmental and in planta regulation of nitrate influx.Ann Bot. 2014 May;113(6):991-1005. doi: 10.1093/aob/mcu021. Epub 2014 Mar 16. Ann Bot. 2014. PMID: 24638820 Free PMC article.
-
A NIGT1-centred transcriptional cascade regulates nitrate signalling and incorporates phosphorus starvation signals in Arabidopsis.Nat Commun. 2018 Apr 10;9(1):1376. doi: 10.1038/s41467-018-03832-6. Nat Commun. 2018. PMID: 29636481 Free PMC article.
-
The Calcium Ion Is a Second Messenger in the Nitrate Signaling Pathway of Arabidopsis.Plant Physiol. 2015 Oct;169(2):1397-404. doi: 10.1104/pp.15.00961. Epub 2015 Aug 24. Plant Physiol. 2015. PMID: 26304850 Free PMC article.
References
-
- Alboresi A, Gestin C, Leydecker MT, Bedu M, Meyer C, Truong HN (2005) Nitrate, a signal relieving seed dormancy in Arabidopsis. Plant Cell Environ 28 500–512 - PubMed
-
- Castaings L, Camargo A, Pocholle D, Gaudon V, Texier Y, Boutet-Mercey S, Taconnat L, Renou JP, Daniel-Vedele F, Fernandez E, et al (2009) The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J 57 426–435 - PubMed
-
- Crawford NM, Glass ADM (1998) Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci 3 389–395
-
- Filleur S, Walch-Liu P, Gan Y, Forde BG (2005) Nitrate and glutamate sensing by plant roots. Biochem Soc Trans 33 283–286 - PubMed
-
- Forde BG (2000) Nitrate transporters in plants: structure, function and regulation. Biochim Biophys Acta 1465 219–235 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

