Multiple roles for MRE11 at uncapped telomeres
- PMID: 19633651
- PMCID: PMC2760383
- DOI: 10.1038/nature08196
Multiple roles for MRE11 at uncapped telomeres
Abstract
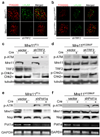
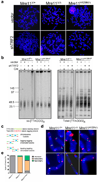
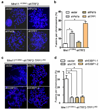
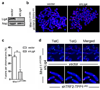
Progressive telomere attrition or uncapping of the shelterin complex elicits a DNA damage response as a result of a cell's inability to distinguish dysfunctional telomeric ends from DNA double-strand breaks. Telomere deprotection activates both ataxia telangiectasia mutated (ATM) and telangiectasia and Rad3-related (ATR) kinase-dependent DNA damage response pathways, and promotes efficient non-homologous end-joining (NHEJ) of dysfunctional telomeres. The mammalian MRE11-RAD50-NBS1 (MRN; NBS1 is also known as NBN) complex interacts with ATM to sense chromosomal double-strand breaks and coordinate global DNA damage responses. Although the MRN complex accumulates at dysfunctional telomeres, it is not known whether mammalian MRN promotes repair at these sites. Here we address this question by using mouse alleles that either inactivate the entire MRN complex or eliminate only the nuclease activities of MRE11 (ref. 8). We show that cells lacking MRN do not activate ATM when telomeric repeat binding factor 2 (TRF2) is removed from telomeres, and ligase 4 (LIG4)-dependent chromosome end-to-end fusions are markedly reduced. Residual chromatid fusions involve only telomeres generated by leading strand synthesis. Notably, although cells deficient for MRE11 nuclease activity efficiently activate ATM and recruit 53BP1 (also known as TP53BP1) to deprotected telomeres, the 3' telomeric overhang persists to prevent NHEJ-mediated chromosomal fusions. Removal of shelterin proteins that protect the 3' overhang in the setting of MRE11 nuclease deficiency restores LIG4-dependent chromosome fusions. Our data indicate a critical role for the MRN complex in sensing dysfunctional telomeres, and show that in the absence of TRF2, MRE11 nuclease activity removes the 3' telomeric overhang to promote chromosome fusions. MRE11 can also protect newly replicated leading strand telomeres from NHEJ by promoting 5' strand resection to generate POT1a-TPP1-bound 3' overhangs.
Figures
References
-
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. - PubMed
-
- van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. - PubMed
-
- Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nature Cell Biol. 2005;7:712–718. - PubMed
-
- Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

