Programming cells by multiplex genome engineering and accelerated evolution
- PMID: 19633652
- PMCID: PMC4590770
- DOI: 10.1038/nature08187
Programming cells by multiplex genome engineering and accelerated evolution
Abstract
The breadth of genomic diversity found among organisms in nature allows populations to adapt to diverse environments. However, genomic diversity is difficult to generate in the laboratory and new phenotypes do not easily arise on practical timescales. Although in vitro and directed evolution methods have created genetic variants with usefully altered phenotypes, these methods are limited to laborious and serial manipulation of single genes and are not used for parallel and continuous directed evolution of gene networks or genomes. Here, we describe multiplex automated genome engineering (MAGE) for large-scale programming and evolution of cells. MAGE simultaneously targets many locations on the chromosome for modification in a single cell or across a population of cells, thus producing combinatorial genomic diversity. Because the process is cyclical and scalable, we constructed prototype devices that automate the MAGE technology to facilitate rapid and continuous generation of a diverse set of genetic changes (mismatches, insertions, deletions). We applied MAGE to optimize the 1-deoxy-D-xylulose-5-phosphate (DXP) biosynthesis pathway in Escherichia coli to overproduce the industrially important isoprenoid lycopene. Twenty-four genetic components in the DXP pathway were modified simultaneously using a complex pool of synthetic DNA, creating over 4.3 billion combinatorial genomic variants per day. We isolated variants with more than fivefold increase in lycopene production within 3 days, a significant improvement over existing metabolic engineering techniques. Our multiplex approach embraces engineering in the context of evolution by expediting the design and evolution of organisms with new and improved properties.
Figures
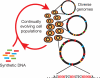

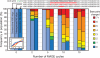

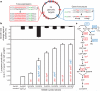
References
-
- Venter JC, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. - PubMed
-
- Tringe SG, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. - PubMed
-
- Elena SF, Lenski RE. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nature Rev. Genet. 2003;4:457–469. - PubMed
-
- Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. - PubMed
-
- Crameri A, Raillard S-A, Bermudez E, Stemmer WP. C. DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature. 1998;391:288–291. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

