Inhibition of a viral enzyme by a small-molecule dimer disruptor
- PMID: 19633659
- PMCID: PMC2752665
- DOI: 10.1038/nchembio.192
Inhibition of a viral enzyme by a small-molecule dimer disruptor
Abstract
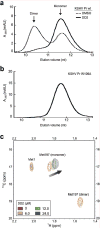
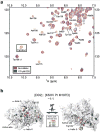
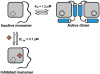
We identified small-molecule dimer disruptors that inhibit an essential dimeric protease of human Kaposi's sarcoma-associated herpesvirus (KSHV) by screening an alpha-helical mimetic library. Next, we synthesized a second generation of low-micromolar inhibitors with improved potency and solubility. Complementary methods including size exclusion chromatography and 1H-13C HSQC titration using selectively labeled 13C-Met samples revealed that monomeric protease is enriched in the presence of inhibitor. 1H-15N HSQC titration studies mapped the inhibitor binding site to the dimer interface, and mutagenesis studies targeting this region were consistent with a mechanism where inhibitor binding prevents dimerization through the conformational selection of a dynamic intermediate. These results validate the interface of herpesvirus proteases and other similar oligomeric interactions as suitable targets for the development of small-molecule inhibitors.
Conflict of interest statement
Figures


Comment in
-
Protease dimer formation disrupted.Nat Chem Biol. 2009 Sep;5(9):607-8. doi: 10.1038/nchembio0909-607. Nat Chem Biol. 2009. PMID: 19690531
References
-
- Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–30. - PubMed
-
- Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285:2177–98. - PubMed
-
- Berg T. Modulation of protein-protein interactions with small organic molecules. Angew Chem Int Ed Engl. 2003;42:2462–81. - PubMed
-
- Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–9. - PubMed
MeSH terms
Substances
Associated data
- PubChem-Substance/81055042
- PubChem-Substance/81055043
- PubChem-Substance/81067414
- PubChem-Substance/81067415
- PubChem-Substance/81067416
- PubChem-Substance/81067417
- PubChem-Substance/81067418
- PubChem-Substance/81067419
- PubChem-Substance/81067420
- PubChem-Substance/81067421
- PubChem-Substance/81067422
- PubChem-Substance/81067423
- PubChem-Substance/81067424
- PubChem-Substance/81067425
- PubChem-Substance/81067426
- PubChem-Substance/81067427
- PubChem-Substance/81067428
- PubChem-Substance/81067429
- PubChem-Substance/81067430
- PubChem-Substance/81067431
- PubChem-Substance/81067432
- PubChem-Substance/81067433
- PubChem-Substance/81067434
- PubChem-Substance/81067435
- PubChem-Substance/81067436
- PubChem-Substance/81067437
Grants and funding
LinkOut - more resources
Full Text Sources
Chemical Information
Miscellaneous

