Role of mammalian Mre11 in classical and alternative nonhomologous end joining
- PMID: 19633669
- PMCID: PMC2730592
- DOI: 10.1038/nsmb.1640
Role of mammalian Mre11 in classical and alternative nonhomologous end joining
Abstract
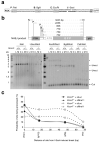
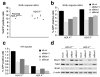
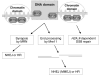
The mammalian Mre11-Rad50-Nbs1 (MRN) complex coordinates double-strand break signaling with repair by homologous recombination and is associated with the H2A.X chromatin response to double-strand breaks, but its role in nonhomologous end joining (NHEJ) is less clear. Here we show that Mre11 promotes efficient NHEJ in both wild-type and Xrcc4(-/-) mouse embryonic stem cells. Depletion of Mre11 reduces the use of microhomology during NHEJ in Xrcc4(+/+) cells and suppresses end resection in Xrcc4(-/-) cells, revealing specific roles for Mre11 in both classical and alternative NHEJ. The NHEJ function of Mre11 is independent of H2A.X. We propose a model in which both enzymatic and scaffolding functions of Mre11 cooperate to support mammalian NHEJ.
Figures

Comment in
-
Mre11: roles in DNA repair beyond homologous recombination.Nat Struct Mol Biol. 2009 Aug;16(8):798-800. doi: 10.1038/nsmb0809-798. Nat Struct Mol Biol. 2009. PMID: 19654615 No abstract available.
References
-
- Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85:509–20. - PubMed
-
- Berkovich E, Monnat RJ, Jr., Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9:683–90. - PubMed
-
- Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–4. - PubMed
-
- Stucki M, Jackson SP. gammaH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair (Amst) 2006;5:534–43. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

