H3.3/H2A.Z double variant-containing nucleosomes mark 'nucleosome-free regions' of active promoters and other regulatory regions
- PMID: 19633671
- PMCID: PMC3125718
- DOI: 10.1038/ng.409
H3.3/H2A.Z double variant-containing nucleosomes mark 'nucleosome-free regions' of active promoters and other regulatory regions
Abstract
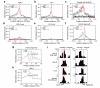
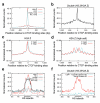
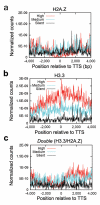
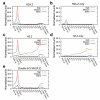
To understand how chromatin structure is organized by different histone variants, we have measured the genome-wide distribution of nucleosome core particles (NCPs) containing the histone variants H3.3 and H2A.Z in human cells. We find that a special class of NCPs containing both variants is enriched at 'nucleosome-free regions' of active promoters, enhancers and insulator regions. We show that preparative methods used previously in studying nucleosome structure result in the loss of these unstable double-variant NCPs. It seems likely that this instability facilitates the access of transcription factors to promoters and other regulatory sites in vivo. Other combinations of variants have different distributions, consistent with distinct roles for histone variants in the modulation of gene expression.
Figures
Comment in
-
Labile H3.3+H2A.Z nucleosomes mark 'nucleosome-free regions'.Nat Genet. 2009 Aug;41(8):865-6. doi: 10.1038/ng0809-865. Nat Genet. 2009. PMID: 19639024 No abstract available.
References
-
- Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet. 2005;37:1090–7. - PubMed
-
- Mito Y, Henikoff JG, Henikoff S. Histone replacement marks the boundaries of cis-regulatory domains. Science. 2007;315:1408–11. - PubMed
-
- Albert I, et al. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–6. - PubMed
-
- Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

