The HIF-1 hypoxia-inducible factor modulates lifespan in C. elegans
- PMID: 19633713
- PMCID: PMC2711329
- DOI: 10.1371/journal.pone.0006348
The HIF-1 hypoxia-inducible factor modulates lifespan in C. elegans
Abstract
During normal development or during disease, animal cells experience hypoxic (low oxygen) conditions, and the hypoxia-inducible factor (HIF) transcription factors implement most of the critical changes in gene expression that enable animals to adapt to this stress. Here, we examine the roles of HIF-1 in post-mitotic aging. We examined the effects of HIF-1 over-expression and of hif-1 loss-of-function mutations on longevity in C. elegans, a powerful genetic system in which adult somatic cells are post-mitotic. We constructed transgenic lines that expressed varying levels of HIF-1 protein and discovered a positive correlation between HIF-1 expression levels and lifespan. The data further showed that HIF-1 acted in parallel to the SKN-1/NRF and DAF-16/FOXO transcription factors to promote longevity. HIF-1 over-expression also conferred increased resistance to heat and oxidative stress. We isolated and characterized additional hif-1 mutations, and we found that each of 3 loss-of-function mutations conferred increased longevity in normal lab culture conditions, but, unlike HIF-1 over-expression, a hif-1 deletion mutation did not extend the lifespan of daf-16 or skn-1 mutants. We conclude that HIF-1 over-expression and hif-1 loss-of-function mutations promote longevity by different pathways. These data establish HIF-1 as one of the key stress-responsive transcription factors that modulate longevity in C. elegans and advance our understanding of the regulatory networks that link oxygen homeostasis and aging.
Conflict of interest statement
Figures
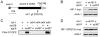





References
-
- Semenza GL. Life with oxygen. Science. 2007;318:62–64. - PubMed
-
- Shen C, Nettleton D, Jiang M, Kim SK, Powell-Coffman JA. Roles of the HIF-1 hypoxia-inducible factor during hypoxia response in Caenorhabditis elegans. J Biol Chem. 2005;280:20580–20588. - PubMed
-
- Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

