Characterization of the PMT gene family in Cryptococcus neoformans
- PMID: 19633715
- PMCID: PMC2711527
- DOI: 10.1371/journal.pone.0006321
Characterization of the PMT gene family in Cryptococcus neoformans
Abstract
Background: Protein-O-mannosyltransferases (Pmt's) catalyze the initial step of protein-O-glycosylation, the addition of mannose residues to serine or threonine residues of target proteins.
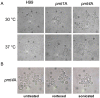
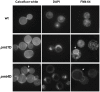
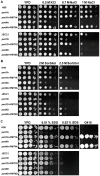
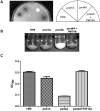
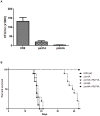
Methodology/principal findings: Based on protein similarities, this highly conserved protein family can be divided into three subfamilies: the Pmt1 sub-family, the Pmt2 sub-family and the Pmt4 sub-family. In contrast to Saccharomyces cerevisiae and Candida albicans, but similar to filamentous fungi, three putative PMT genes (PMT1, PMT2, and PMT4) were identified in the genome of the human fungal pathogen Cryptococcus neoformans. Similar to Schizosaccharomyces pombe and C. albicans, C. neoformans PMT2 is an essential gene. In contrast, the pmt1 and pmt4 single mutants are viable; however, the pmt1/pmt4 deletions are synthetically lethal. Mutation of PMT1 and PMT4 resulted in distinct defects in cell morphology and cell integrity. The pmt1 mutant was more susceptible to SDS medium than wild-type strains and the mutant cells were enlarged. The pmt4 mutant grew poorly on high salt medium and demonstrated abnormal septum formation and defects in cell separation. Interestingly, the pmt1 and pmt4 mutants demonstrated variety-specific differences in the levels of susceptibility to osmotic and cell wall stress. Delayed melanin production in the pmt4 mutant was the only alteration of classical virulence-associated phenotypes. However, the pmt1 and pmt4 mutants showed attenuated virulence in a murine inhalation model of cryptococcosis.
Conclusion/significance: These findings suggest that C. neoformans protein-O-mannosyltransferases play a crucial role in maintaining cell morphology, and that reduced protein-O-glycosylation leads to alterations in stress resistance, cell wall composition, cell integrity, and survival within the host.
Conflict of interest statement
Figures


References
-
- Tanner W, Lehle L. Protein glycosylation in yeast. Biochim Biophys Acta. 1987;906:81–99. - PubMed
-
- Strahl-Bolsinger S, Gentzsch M, Tanner W. Protein O-mannosylation. Biochim Biophys Acta. 1999;1426:297–307. - PubMed
-
- Ernst JF, Prill SK. O-glycosylation. Med Mycol. 2001;39(Suppl 1):67–74. - PubMed
-
- Jurado LA, Coloma A, Cruces J. Identification of a human homolog of the Drosophila rotated abdomen gene (POMT1) encoding a putative protein O-mannosyl-transferase, and assignment to human chromosome 9q34.1. Genomics. 1999;58:171–180. - PubMed
-
- Willer T, Amselgruber W, Deutzmann R, Strahl S. Characterization of POMT2, a novel member of the PMT protein O-mannosyltransferase family specifically localized to the acrosome of mammalian spermatids. Glycobiology. 2002;12:771–783. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

