Evidence of recombination and genetic diversity in human rhinoviruses in children with acute respiratory infection
- PMID: 19633719
- PMCID: PMC2712091
- DOI: 10.1371/journal.pone.0006355
Evidence of recombination and genetic diversity in human rhinoviruses in children with acute respiratory infection
Abstract
Background: Human rhinoviruses (HRVs) are a highly prevalent cause of acute respiratory infection in children. They are classified into at least three species, HRV-A, HRV-B and HRV-C, which are characterized by sequencing the 5' untranslated region (UTR) or the VP4/VP2 region of the genome. Given the increased interest for novel HRV strain identification and their worldwide distribution, we have carried out clinical and molecular diagnosis of HRV strains in a 2-year study of children with acute respiratory infection visiting one district hospital in Shanghai.
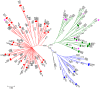
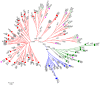
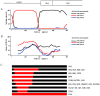
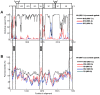
Methodology/findings: We cloned and sequenced a 924-nt fragment that covered part of the 5'UTR and the VP4/VP2 capsid genes. Sixty-four HRV-infected outpatients were diagnosed amongst 827 children with acute low respiratory tract infection. Two samples were co-infected with HRV-A and HRV-B or HRV-C. By comparative analysis of the VP4/VP2 sequences of the 66 HRVs, we showed a high diversity of strains in HRV-A and HRV-B species, and a prevalence of 51.5% of strains that belonged to the recently identified HRV-C species. When analyzing a fragment of the 5' UTR, we characterized at least two subspecies of HRV-C: HRV-Cc, which clustered differently from HRV-A and HRV-B, and HRV-Ca, which resulted from previous recombination in this region with sequences related to HRV-A. The full-length sequence of one strain of each HRV-Ca and HRV-Cc subspecies was obtained for comparative analysis. We confirmed the close relationship of their structural proteins but showed apparent additional recombination events in the 2A gene and 3'UTR of the HRV-Ca strain. Double or triple infections with HRV-C and respiratory syncytial virus and/or bocavirus were diagnosed in 33.3% of the HRV-infected patients, but no correlation with severity of clinical outcome was observed.
Conclusion: Our study showed a high diversity of HRV strains that cause bronchitis and pneumonia in children. A predominance of HRV-C over HRV-A and HRV-B was observed, and two subspecies of HRV-C were identified, the diversity of which seemed to be related to recombination with former HRV-A strains. None of the HRV-C strains appeared to have a higher clinical impact than HRV-A or HRV-B on respiratory compromise.
Conflict of interest statement
Figures

References
-
- Brownlee JW, Turner RB. New developments in the epidemiology and clinical spectrum of rhinovirus infections. Curr Opin Pediatr. 2008;20:67–71. - PubMed
-
- Jacques J, Bouscambert-Duchamp M, Moret H, Carquin J, Brodard V, et al. Association of respiratory picornaviruses with acute bronchiolitis in French infants. J Clin Virol. 2006;35:463–466. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

