Mutational biases and selective forces shaping the structure of Arabidopsis genes
- PMID: 19633720
- PMCID: PMC2712092
- DOI: 10.1371/journal.pone.0006356
Mutational biases and selective forces shaping the structure of Arabidopsis genes
Abstract
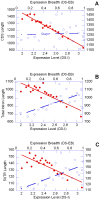
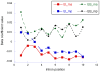
Recently features of gene expression profiles have been associated with structural parameters of gene sequences in organisms representing a diverse set of taxa. The emerging picture indicates that natural selection, mediated by gene expression profiles, has a significant role in determining genic structures. However the current situation is less clear in plants as the available data indicates that the effect of natural selection mediated by gene expression is very weak. Moreover, the direction of the patterns in plants appears to contradict those observed in animal genomes. In the present work we analized expression data for >18000 Arabidopsis genes retrieved from public datasets obtained with different technologies (MPSS and high density chip arrays) and compared them with gene parameters. Our results show that the impact of natural selection mediated by expression on genes sequences is significant and distinguishable from the effects of regional mutational biases. In addition, we provide evidence that the level and the breadth of gene expression are related in opposite ways to many structural parameters of gene sequences. Higher levels of expression abundance are associated with smaller transcripts, consistent with the need to reduce costs of both transcription and translation. Expression breadth, however, shows a contrasting pattern, i.e. longer genes have higher breadth of expression, possibly to ensure those structural features associated with gene plasticity. Based on these results, we propose that the specific balance between these two selective forces play a significant role in shaping the structure of Arabidopsis genes.
Conflict of interest statement
Figures

References
-
- Castillo-Davis CI, Mekhedov SL, Hartl DL, Koonin EV, Kondrashov FA. Selection for short introns in highly expressed genes. Nat Genet. 2002;31:415–418. - PubMed
-
- Vinogradov AE. Compactness of human housekeeping genes: selection for economy or genomic design? Trends Genet. 2004;20:248–253. - PubMed
-
- Eisenberg E, Levanon EY. Human housekeeping genes are compact. Trends Genet. 2003;19:362–365. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

