New insights into short-chain prenyltransferases: structural features, evolutionary history and potential for selective inhibition
- PMID: 19633972
- PMCID: PMC11115643
- DOI: 10.1007/s00018-009-0100-9
New insights into short-chain prenyltransferases: structural features, evolutionary history and potential for selective inhibition
Abstract
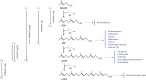
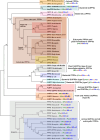
Isoprenoids form an extensive group of natural products involved in a number of important biological processes. Their biosynthesis proceeds through sequential 1'-4 condensations of isopentenyl diphosphate (C5) with an allylic acceptor, the first of which is dimethylallyl diphosphate (C5). The reactions leading to the production of geranyl diphosphate (C10), farnesyl diphosphate (C15) and geranylgeranyl diphosphate (C20), which are the precursors of mono-, sesqui- and diterpenes, respectively, are catalyzed by a group of highly conserved enzymes known as short-chain isoprenyl diphosphate synthases, or prenyltransferases. In recent years, the sequences of many new prenyltransferases have become available, including those of several plant and animal geranyl diphosphate synthases, revealing novel mechanisms of product chain-length selectivity and an intricate evolutionary path from a putative common ancestor. Finally, there is considerable interest in designing inhibitors specific to short-chain prenyltransferases, for the purpose of developing new drugs or pesticides that target the isoprenoid biosynthetic pathway.
Figures
References
-
- Ogura K, Toyama T, Sagami H. Polyprenyl diphopshate synthases. In: Bittman R, editor. Subcellular biochemistry. New York: Plenum; 1997. pp. 57–87. - PubMed
-
- Ko TP, Chen YK, Robinson H, Tsai PC, Gao YG, Chen APC, Wang AHJ, Liang PH. Mechanism of product chain length determination and the role of a flexible loop in Escherichia coli undecaprenyl-pyrophosphate synthase catalysis. J Biol Chem. 2001;276:47474–47482. doi: 10.1074/jbc.M106747200. - DOI - PubMed
-
- Croteau R, Purkett PT. Geranyl pyrophosphate synthase: characterization of the enzyme and evidence that this chain-length specific prenyltransferase is associated with monoterpene biosynthesis in sage (Salvia officinalis) Arch Biochem Biophys. 1989;271:524–535. doi: 10.1016/0003-9861(89)90304-4. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

