Critical factors determining dimerization of human antizyme inhibitor
- PMID: 19635796
- PMCID: PMC2785365
- DOI: 10.1074/jbc.M109.007807
Critical factors determining dimerization of human antizyme inhibitor
Abstract
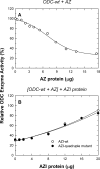
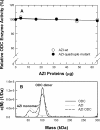
Ornithine decarboxylase (ODC) is the first enzyme involved in polyamine biosynthesis, and it catalyzes the decarboxylation of ornithine to putrescine. ODC is a dimeric enzyme, whereas antizyme inhibitor (AZI), a positive regulator of ODC that is homologous to ODC, exists predominantly as a monomer and lacks decarboxylase activity. The goal of this paper was to identify the essential amino acid residues that determine the dimerization of AZI. The nonconserved amino acid residues in the putative dimer interface of AZI (Ser-277, Ser-331, Glu-332, and Asp-389) were substituted with the corresponding residues in the putative dimer interface of ODC (Arg-277, Tyr-331, Asp-332, and Tyr-389, respectively). Analytical ultracentrifugation analysis was used to determine the size distribution of these AZI mutants. The size-distribution analysis data suggest that residue 331 may play a major role in the dimerization of AZI. Mutating Ser-331 to Tyr in AZI (AZI-S331Y) caused a shift from a monomer configuration to a dimer. Furthermore, in comparison with the single mutant AZI-S331Y, the AZI-S331Y/D389Y double mutant displayed a further reduction in the monomer-dimer K(d), suggesting that residue 389 is also crucial for AZI dimerization. Analysis of the triple mutant AZI-S331Y/D389Y/S277R showed that it formed a stable dimer (K(d) value = 1.3 microm). Finally, a quadruple mutant, S331Y/D389Y/S277R/E332D, behaved as a dimer with a K(d) value of approximately 0.1 microm, which is very close to that of the human ODC enzyme. The quadruple mutant, although forming a dimer, could still be disrupted by antizyme (AZ), further forming a heterodimer, and it could rescue the AZ-inhibited ODC activity, suggesting that the AZ-binding ability of the AZI dimer was retained.
Figures






Similar articles
-
Minimal antizyme peptide fully functioning in the binding and inhibition of ornithine decarboxylase and antizyme inhibitor.PLoS One. 2011;6(9):e24366. doi: 10.1371/journal.pone.0024366. Epub 2011 Sep 9. PLoS One. 2011. PMID: 21931692 Free PMC article.
-
Critical factors governing the difference in antizyme-binding affinities between human ornithine decarboxylase and antizyme inhibitor.PLoS One. 2011 Apr 28;6(4):e19253. doi: 10.1371/journal.pone.0019253. PLoS One. 2011. PMID: 21552531 Free PMC article.
-
Crystallographic and biochemical studies revealing the structural basis for antizyme inhibitor function.Protein Sci. 2008 May;17(5):793-802. doi: 10.1110/ps.073427208. Epub 2008 Mar 27. Protein Sci. 2008. PMID: 18369191 Free PMC article.
-
The antizyme family for regulating polyamines.J Biol Chem. 2018 Nov 30;293(48):18730-18735. doi: 10.1074/jbc.TM118.003339. Epub 2018 Oct 24. J Biol Chem. 2018. PMID: 30355739 Free PMC article. Review.
-
Regulation of cellular polyamine levels and cellular proliferation by antizyme and antizyme inhibitor.Essays Biochem. 2009 Nov 4;46:47-61. doi: 10.1042/bse0460004. Essays Biochem. 2009. PMID: 20095969 Review.
Cited by
-
Recurrent emergence of catalytically inactive ornithine decarboxylase homologous forms that likely have regulatory function.J Mol Evol. 2010 Mar;70(3):289-302. doi: 10.1007/s00239-010-9331-5. Epub 2010 Mar 9. J Mol Evol. 2010. PMID: 20217058
-
Multifaceted interactions and regulation between antizyme and its interacting proteins cyclin D1, ornithine decarboxylase and antizyme inhibitor.Oncotarget. 2015 Sep 15;6(27):23917-29. doi: 10.18632/oncotarget.4469. Oncotarget. 2015. PMID: 26172301 Free PMC article.
-
Structural and degradative aspects of ornithine decarboxylase antizyme inhibitor 2.FEBS Open Bio. 2014 Jun 2;4:510-21. doi: 10.1016/j.fob.2014.05.004. eCollection 2014. FEBS Open Bio. 2014. PMID: 24967154 Free PMC article.
-
Minimal antizyme peptide fully functioning in the binding and inhibition of ornithine decarboxylase and antizyme inhibitor.PLoS One. 2011;6(9):e24366. doi: 10.1371/journal.pone.0024366. Epub 2011 Sep 9. PLoS One. 2011. PMID: 21931692 Free PMC article.
-
Novel interaction of ornithine decarboxylase with sepiapterin reductase regulates neuroblastoma cell proliferation.J Mol Biol. 2014 Jan 23;426(2):332-46. doi: 10.1016/j.jmb.2013.09.037. Epub 2013 Oct 1. J Mol Biol. 2014. PMID: 24096079 Free PMC article.
References
-
- Gerner E. W., Meyskens F. L., Jr. (2004) Nat. Rev. Cancer 4, 781–792 - PubMed
-
- Pegg A. E., McCann P. P. (1982) Am. J. Physiol. 243, C212–C221 - PubMed
-
- Tabor C. W., Tabor H. (1984) Annu. Rev. Biochem. 53, 749–790 - PubMed
-
- Mangold U. (2005) IUBMB Life 57, 671–676 - PubMed
-
- Pegg A. E. (2006) J. Biol. Chem. 281, 14529–14532 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

