Structural basis for the mechanism of respiratory complex I
- PMID: 19635800
- PMCID: PMC2785608
- DOI: 10.1074/jbc.M109.032144
Structural basis for the mechanism of respiratory complex I
Abstract
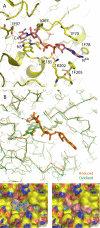
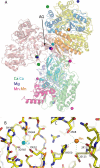
Complex I plays a central role in cellular energy production, coupling electron transfer between NADH and quinone to proton translocation. The mechanism of this highly efficient enzyme is currently unknown. Mitochondrial complex I is a major source of reactive oxygen species, which may be one of the causes of aging. Dysfunction of complex I is implicated in many human neurodegenerative diseases. We have determined several x-ray structures of the oxidized and reduced hydrophilic domain of complex I from Thermus thermophilus at up to 3.1 A resolution. The structures reveal the mode of interaction of complex I with NADH, explaining known kinetic data and providing implications for the mechanism of reactive oxygen species production at the flavin site of complex I. Bound metals were identified in the channel at the interface with the frataxin-like subunit Nqo15, indicating possible iron-binding sites. Conformational changes upon reduction of the complex involve adjustments in the nucleotide-binding pocket, as well as small but significant shifts of several alpha-helices at the interface with the membrane domain. These shifts are likely to be driven by the reduction of nearby iron-sulfur clusters N2 and N6a/b. Cluster N2 is the electron donor to quinone and is coordinated by unique motif involving two consecutive (tandem) cysteines. An unprecedented "on/off switch" (disconnection) of coordinating bonds between the tandem cysteines and this cluster was observed upon reduction. Comparison of the structures suggests a novel mechanism of coupling between electron transfer and proton translocation, combining conformational changes and protonation/deprotonation of tandem cysteines.
Figures



References
-
- Walker J. E. (1992) Q. Rev. Biophys. 25, 253–324 - PubMed
-
- Yagi T., Matsuno-Yagi A. (2003) Biochemistry 42, 2266–2274 - PubMed
-
- Carroll J., Fearnley I. M., Skehel J. M., Shannon R. J., Hirst J., Walker J. E. (2006) J. Biol. Chem. 281, 32724–32727 - PubMed
-
- Sazanov L. A. (2007) Biochemistry 46, 2275–2288 - PubMed
-
- Schapira A. H. (1998) Biochim. Biophys. Acta 1364, 261–270 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

