The Saccharomyces cerevisiae Rad6 postreplication repair and Siz1/Srs2 homologous recombination-inhibiting pathways process DNA damage that arises in asf1 mutants
- PMID: 19635810
- PMCID: PMC2747975
- DOI: 10.1128/MCB.00894-09
The Saccharomyces cerevisiae Rad6 postreplication repair and Siz1/Srs2 homologous recombination-inhibiting pathways process DNA damage that arises in asf1 mutants
Abstract
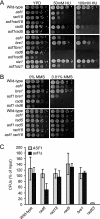
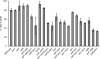
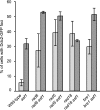
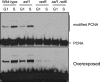
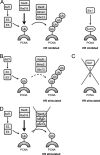
The Asf1 and Rad6 pathways have been implicated in a number of common processes such as suppression of gross chromosomal rearrangements (GCRs), DNA repair, modification of chromatin, and proper checkpoint functions. We examined the relationship between Asf1 and different gene products implicated in postreplication repair (PRR) pathways in the suppression of GCRs, checkpoint function, sensitivity to hydroxyurea (HU) and methyl methanesulfonate (MMS), and ubiquitination of proliferating cell nuclear antigen (PCNA). We found that defects in Rad6 PRR pathway and Siz1/Srs2 homologous recombination suppression (HRS) pathway genes suppressed the increased GCR rates seen in asf1 mutants, which was independent of translesion bypass polymerases but showed an increased dependency on Dun1. Combining an asf1 deletion with different PRR mutations resulted in a synergistic increase in sensitivity to chronic HU and MMS treatment; however, these double mutants were not checkpoint defective, since they were capable of recovering from acute treatment with HU. Interestingly, we found that Asf1 and Rad6 cooperate in ubiquitination of PCNA, indicating that Rad6 and Asf1 function in parallel pathways that ubiquitinate PCNA. Our results show that ASF1 probably contributes to the maintenance of genome stability through multiple mechanisms, some of which involve the PRR and HRS pathways.
Figures

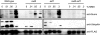
Similar articles
-
Suppression of genetic defects within the RAD6 pathway by srs2 is specific for error-free post-replication repair but not for damage-induced mutagenesis.Nucleic Acids Res. 2002 Feb 1;30(3):732-9. doi: 10.1093/nar/30.3.732. Nucleic Acids Res. 2002. PMID: 11809886 Free PMC article.
-
Checkpoint functions are required for normal S-phase progression in Saccharomyces cerevisiae RCAF- and CAF-I-defective mutants.Proc Natl Acad Sci U S A. 2006 Mar 7;103(10):3710-5. doi: 10.1073/pnas.0511102103. Epub 2006 Feb 24. Proc Natl Acad Sci U S A. 2006. PMID: 16501045 Free PMC article.
-
Srs2 plays a critical role in reversible G2 arrest upon chronic and low doses of UV irradiation via two distinct homologous recombination-dependent mechanisms in postreplication repair-deficient cells.Mol Cell Biol. 2010 Oct;30(20):4840-50. doi: 10.1128/MCB.00453-10. Epub 2010 Aug 16. Mol Cell Biol. 2010. PMID: 20713444 Free PMC article.
-
Error-free DNA-damage tolerance in Saccharomyces cerevisiae.Mutat Res Rev Mutat Res. 2015 Apr-Jun;764:43-50. doi: 10.1016/j.mrrev.2015.02.001. Epub 2015 Feb 16. Mutat Res Rev Mutat Res. 2015. PMID: 26041265 Review.
-
DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae.Mutat Res. 2001 Aug 9;486(3):167-84. doi: 10.1016/s0921-8777(01)00091-x. Mutat Res. 2001. PMID: 11459630 Review.
Cited by
-
SUMO Wrestles with Recombination.Biomolecules. 2012 Jul 25;2(3):350-75. doi: 10.3390/biom2030350. Biomolecules. 2012. PMID: 24970142 Free PMC article.
-
Sumo and the cellular stress response.Cell Div. 2015 Jun 20;10:4. doi: 10.1186/s13008-015-0010-1. eCollection 2015. Cell Div. 2015. PMID: 26101541 Free PMC article. Review.
-
Damage-specific modification of PCNA.Cell Cycle. 2010 Sep 15;9(18):3674-9. doi: 10.4161/cc.9.18.13121. Epub 2010 Sep 21. Cell Cycle. 2010. PMID: 20930510 Free PMC article.
-
A genetic and structural study of genome rearrangements mediated by high copy repeat Ty1 elements.PLoS Genet. 2011 May;7(5):e1002089. doi: 10.1371/journal.pgen.1002089. Epub 2011 May 26. PLoS Genet. 2011. PMID: 21637792 Free PMC article.
-
A saccharomyces cerevisiae RNase H2 interaction network functions to suppress genome instability.Mol Cell Biol. 2014 Apr;34(8):1521-34. doi: 10.1128/MCB.00960-13. Epub 2014 Feb 18. Mol Cell Biol. 2014. PMID: 24550002 Free PMC article.
References
-
- Adkins, M. W., S. R. Howar, and J. K. Tyler. 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14:657-666. - PubMed
-
- Adkins, M. W., and J. K. Tyler. 2004. The histone chaperone Asf1p mediates global chromatin disassembly in vivo. J. Biol. Chem. 279:52069-52074. - PubMed
-
- Bailly, V., J. Lamb, P. Sung, S. Prakash, and L. Prakash. 1994. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 8:811-820. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
