The polymerase eta translesion synthesis DNA polymerase acts independently of the mismatch repair system to limit mutagenesis caused by 7,8-dihydro-8-oxoguanine in yeast
- PMID: 19635811
- PMCID: PMC2747974
- DOI: 10.1128/MCB.00422-09
The polymerase eta translesion synthesis DNA polymerase acts independently of the mismatch repair system to limit mutagenesis caused by 7,8-dihydro-8-oxoguanine in yeast
Abstract
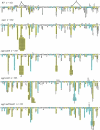
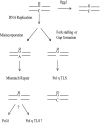
Reactive oxygen species are ubiquitous mutagens that have been linked to both disease and aging. The most studied oxidative lesion is 7,8-dihydro-8-oxoguanine (GO), which is often miscoded during DNA replication, resulting specifically in GC --> TA transversions. In yeast, the mismatch repair (MMR) system repairs GO.A mismatches generated during DNA replication, and the polymerase eta (Poleta) translesion synthesis DNA polymerase additionally promotes error-free bypass of GO lesions. It has been suggested that Poleta limits GO-associated mutagenesis exclusively through its participation in the filling of MMR-generated gaps that contain GO lesions. In the experiments reported here, the SUP4-o forward-mutation assay was used to monitor GC --> TA mutation rates in strains defective in MMR (Msh2 or Msh6) and/or in Poleta activity. The results clearly demonstrate that Poleta can function independently of the MMR system to prevent GO-associated mutations, presumably through preferential insertion of cytosine opposite replication-blocking GO lesions. Furthermore, the Poleta-dependent bypass of GO lesions is more efficient on the lagging strand of replication and requires an interaction with proliferating cell nuclear antigen. These studies establish a new paradigm for the prevention of GO-associated mutagenesis in eukaryotes.
Figures
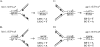

Similar articles
-
The post-replication repair RAD18 and RAD6 genes are involved in the prevention of spontaneous mutations caused by 7,8-dihydro-8-oxoguanine in Saccharomyces cerevisiae.Nucleic Acids Res. 2004 Sep 23;32(17):5003-10. doi: 10.1093/nar/gkh831. Print 2004. Nucleic Acids Res. 2004. PMID: 15388802 Free PMC article.
-
PCNA monoubiquitylation and DNA polymerase eta ubiquitin-binding domain are required to prevent 8-oxoguanine-induced mutagenesis in Saccharomyces cerevisiae.Nucleic Acids Res. 2009 May;37(8):2549-59. doi: 10.1093/nar/gkp105. Epub 2009 Mar 5. Nucleic Acids Res. 2009. PMID: 19264809 Free PMC article.
-
Role of DNA polymerase eta in the bypass of abasic sites in yeast cells.Nucleic Acids Res. 2004 Jul 29;32(13):3984-94. doi: 10.1093/nar/gkh710. Print 2004. Nucleic Acids Res. 2004. PMID: 15284331 Free PMC article.
-
Repair of 8-oxoguanine in Saccharomyces cerevisiae: interplay of DNA repair and replication mechanisms.Free Radic Biol Med. 2002 Jun 15;32(12):1244-53. doi: 10.1016/s0891-5849(02)00822-5. Free Radic Biol Med. 2002. PMID: 12057762 Review.
-
Exonuclease 1-dependent and independent mismatch repair.DNA Repair (Amst). 2015 Aug;32:24-32. doi: 10.1016/j.dnarep.2015.04.010. Epub 2015 Apr 30. DNA Repair (Amst). 2015. PMID: 25956862 Free PMC article. Review.
Cited by
-
DNA polymerase η contributes to genome-wide lagging strand synthesis.Nucleic Acids Res. 2019 Mar 18;47(5):2425-2435. doi: 10.1093/nar/gky1291. Nucleic Acids Res. 2019. PMID: 30597049 Free PMC article.
-
DNA mismatch repair system: repercussions in cellular homeostasis and relationship with aging.Oxid Med Cell Longev. 2012;2012:728430. doi: 10.1155/2012/728430. Epub 2012 Nov 8. Oxid Med Cell Longev. 2012. PMID: 23213348 Free PMC article. Review.
-
DNA repair mechanisms and the bypass of DNA damage in Saccharomyces cerevisiae.Genetics. 2013 Apr;193(4):1025-64. doi: 10.1534/genetics.112.145219. Genetics. 2013. PMID: 23547164 Free PMC article. Review.
-
Global analysis of mutations driving microevolution of a heterozygous diploid fungal pathogen.Proc Natl Acad Sci U S A. 2018 Sep 11;115(37):E8688-E8697. doi: 10.1073/pnas.1806002115. Epub 2018 Aug 27. Proc Natl Acad Sci U S A. 2018. PMID: 30150418 Free PMC article.
-
Genotoxicity of tri- and hexavalent chromium compounds in vivo and their modes of action on DNA damage in vitro.PLoS One. 2014 Aug 11;9(8):e103194. doi: 10.1371/journal.pone.0103194. eCollection 2014. PLoS One. 2014. PMID: 25111056 Free PMC article.
References
-
- Abdulovic, A. L., B. K. Minesinger, and S. Jinks-Robertson. 2007. Identification of a strand-related bias in the PCNA-mediated bypass of spontaneous lesions by yeast Polη. DNA Repair (Amsterdam) 6:1307-1318. - PubMed
-
- Bahler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. - PubMed
-
- Bebenek, K., J. C. Boyer, and T. A. Kunkel. 1999. The base substitution fidelity of HIV-1 reverse transcriptase on DNA and RNA templates probed with 8-oxo-deoxyguanosine triphosphate. Mutat. Res. 429:149-158. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
