Henipavirus RNA in African bats
- PMID: 19636378
- PMCID: PMC2712088
- DOI: 10.1371/journal.pone.0006367
Henipavirus RNA in African bats
Abstract
Background: Henipaviruses (Hendra and Nipah virus) are highly pathogenic members of the family Paramyxoviridae. Fruit-eating bats of the Pteropus genus have been suggested as their natural reservoir. Human Henipavirus infections have been reported in a region extending from Australia via Malaysia into Bangladesh, compatible with the geographic range of Pteropus. These bats do not occur in continental Africa, but a whole range of other fruit bats is encountered. One of the most abundant is Eidolon helvum, the African Straw-coloured fruit bat.
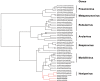
Methodology/principal findings: Feces from E. helvum roosting in an urban setting in Kumasi/Ghana were tested for Henipavirus RNA. Sequences of three novel viruses in phylogenetic relationship to known Henipaviruses were detected. Virus RNA concentrations in feces were low.
Conclusions/significance: The finding of novel putative Henipaviruses outside Australia and Asia contributes a significant extension of the region of potential endemicity of one of the most pathogenic virus genera known in humans.
Conflict of interest statement
Figures

References
-
- Lamb RA, Collins PL, Kolakofsky D, Melero JA, Nagai Y, et al. Family paramyxoviridae. In: Fauquet CM, Mayo J, Maniloff J, Desselberger U, Ball LA, editors. Virus taxonomy: 8th report of the International Committee on Taxonomy of Viruses. San Diego: Elsevier; 2005. pp. 655–668.
-
- Lamb RA, Parks GD. Paramyxoviridae: the viruses and their replication. In: Knipe DM, Griffin DE, Lamb RA, Straus SE, Howley PM, et al., editors. Fields virology. Philadelphia: Lippincott; 2007. pp. 1449–1496.
-
- Wang LF, Chua KB, Yu M, Eaton BT. Genome Diversity of Emerging Paramyxoviruses. Current Genomics. 2003;4:263–273.
-
- Chua KB, Bellini WJ, Rota PA, Harcourt BH, Tamin A, et al. Nipah virus: a recently emergent deadly paramyxovirus. Science. 2000;288:1432–1435. - PubMed
-
- Murray K, Selleck P, Hooper P, Hyatt A, Gould A, et al. A morbillivirus that caused fatal disease in horses and humans. Science. 1995;268:94–97. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

