Dynamics of individual polymers using microfluidic based microcurvilinear flow
- PMID: 19636465
- PMCID: PMC2849989
- DOI: 10.1039/b907860f
Dynamics of individual polymers using microfluidic based microcurvilinear flow
Abstract
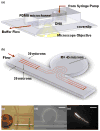
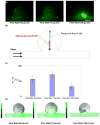
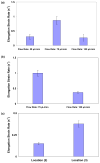
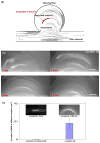
Polymer dynamics play an important role in a diversity of fields including materials science, physics, biology and medicine. The spatiotemporal responses of individual molecules such as biopolymers have been critical to the development of new materials, the expanded understanding of cell structures including cytoskeletal dynamics, and DNA replication. The ability to probe single molecule dynamics however is often limited by the availability of small-scale technologies that can manipulate these systems to uncover highly intricate behaviors. Advances in micro- and nano-scale technologies have simultaneously provided us with valuable tools that can interface with these systems including methods such as microfluidics. Here, we report on the creation of micro-curvilinear flow through a small-scale fluidic approach, which we have been used to impose a flow-based high radial acceleration ( approximately 10(3) g) on individual flexible polymers. We were able to employ this microfluidic-based approach to adjust and control flow velocity and acceleration to observe real-time dynamics of fluorescently labeled lambda-phage DNA molecules in our device. This allowed us to impose mechanical stimulation including stretching and bending on single molecules in localized regimes through a simple and straightforward technology-based method. We found that the flexible DNA molecules exhibited multimodal responses including distinct conformations and controllable curvatures; these characteristics were directly related to both the elongation and bending dynamics dictated by their locations within the curvilinear flow. We analyzed the dynamics of these individual molecules to determine their elongation strain rates and curvatures ( approximately 0.09 microm(-1)) at different locations in this system to probe the individual polymer structural response. These results demonstrate our ability to create high radial acceleration flow and observe real-time dynamic responses applied directly to individual DNA molecules. This approach may also be useful for studying other biologically based polymers including additional nucleic acids, actin filaments, and microtubules and provide a platform to understand the material properties of flexible polymers at a small scale.
Figures

References
-
- Flory P. Statistical Mechanics of Chain Molecules. Interscience; New York: 1969.
-
- de Gennes PG. Scaling Concepts in Polymer Physics. Cornell University Press; Ithaca, NY: 1979.
-
- Doi M, Edwards S. The Theory of Polymer Dynamics. Clarendon; Oxford: 1986.
-
- Fuller GG. Optical Rheometry of Complex Fluids. Oxford University Press; New York: 1995.
-
- Perkins TT, Quake SR, Smith DE, Chu S. Science. 1994;264:822–826. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

