Characterisation of urokinase plasminogen activator receptor variants in human airway and peripheral cells
- PMID: 19638192
- PMCID: PMC2724484
- DOI: 10.1186/1471-2199-10-75
Characterisation of urokinase plasminogen activator receptor variants in human airway and peripheral cells
Abstract
Background: Expression of the urokinase plasminogen activator receptor (UPAR) has been shown to have clinical relevance in various cancers. We have recently identified UPAR as an asthma susceptibility gene and there is evidence to suggest that uPAR may be upregulated in lung diseases such as COPD and asthma. uPAR is a key receptor involved in the formation of the serine protease plasmin by interacting with uPA and has been implicated in many physiological processes including proliferation and migration. The current aim was to determine key regulatory regions and splice variants of UPAR and quantify its expression in primary human tissues and cells (including lung, bronchial epithelium (HBEC), airway smooth muscle (HASM) and peripheral cells).
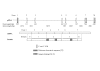
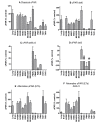
Results: Using Rapid Amplification of cDNA Ends (RACE) a conserved transcription start site (-42 to -77 relative to ATG) was identified and multiple transcription factor binding sites predicted. Seven major splice variants were identified (>5% total expression), including multiple exon deletions and an alternative exon 7b (encoding a truncated, soluble, 229aa protein). Variants were differentially expressed, with a high proportion of E7b usage in lung tissue and structural cells (55-87% of transcripts), whereas classical exon 7 (encoding the GPI-linked protein) was preferentially expressed in peripheral cells (approximately 80% of transcripts), often with exon 6 or 5+6 deletions. Real-time PCR confirmed expression of uPAR mRNA in lung, as well as airway and peripheral cell types with ~50-100 fold greater expression in peripheral cells versus airway cells and confirmed RACE data. Protein analysis confirmed expression of multiple different forms of uPAR in the same cells as well as expression of soluble uPAR in cell supernatants. The pattern of expression did not directly reflect that seen at the mRNA level, indicating that post-translational mechanisms of regulation may also play an important role.
Conclusion: We have identified multiple uPAR isoforms in the lung and immune cells and shown that expression is cell specific. These data provide a novel mechanism for uPAR regulation, as different exon splicing may determine uPAR function e.g. alternative E7b results in a soluble isoform due to the loss of the GPI anchor and exon deletions may affect uPA (ligand) and/or integrin binding and therefore influence downstream pathways. Expression of different isoforms within the lung should be taken into consideration in studies of uPAR in respiratory disease.
Figures








Similar articles
-
Cigarette Smoke and the Induction of Urokinase Plasminogen Activator Receptor In Vivo: Selective Contribution of Isoforms to Bronchial Epithelial Phenotype.Am J Respir Cell Mol Biol. 2015 Aug;53(2):174-83. doi: 10.1165/rcmb.2014-0296OC. Am J Respir Cell Mol Biol. 2015. PMID: 25490122
-
Airway and peripheral urokinase plasminogen activator receptor is elevated in asthma, and identifies a severe, nonatopic subset of patients.Allergy. 2017 Mar;72(3):473-482. doi: 10.1111/all.13046. Epub 2016 Oct 5. Allergy. 2017. PMID: 27624865
-
uPAR regulates bronchial epithelial repair in vitro and is elevated in asthmatic epithelium.Thorax. 2012 Jun;67(6):477-87. doi: 10.1136/thoraxjnl-2011-200508. Epub 2011 Dec 3. Thorax. 2012. PMID: 22139533 Free PMC article.
-
Structure, function and expression on blood and bone marrow cells of the urokinase-type plasminogen activator receptor, uPAR.Stem Cells. 1997;15(6):398-408. doi: 10.1002/stem.150398. Stem Cells. 1997. PMID: 9402652 Review.
-
The role and regulation of urokinase-type plasminogen activator receptor gene expression in cancer invasion and metastasis.Med Res Rev. 2001 Mar;21(2):146-70. doi: 10.1002/1098-1128(200103)21:2<146::aid-med1004>3.0.co;2-b. Med Res Rev. 2001. PMID: 11223863 Review.
Cited by
-
Genome-wide protein QTL mapping identifies human plasma kallikrein as a post-translational regulator of serum uPAR levels.FASEB J. 2014 Feb;28(2):923-34. doi: 10.1096/fj.13-240879. Epub 2013 Nov 18. FASEB J. 2014. PMID: 24249636 Free PMC article.
-
PLAUR polymorphisms and lung function in UK smokers.BMC Med Genet. 2009 Oct 31;10:112. doi: 10.1186/1471-2350-10-112. BMC Med Genet. 2009. PMID: 19878584 Free PMC article.
-
Elevated PLAUR is observed in the airway epithelium of asthma patients and blocking improves barrier integrity.Clin Transl Allergy. 2023 Oct;13(10):e12293. doi: 10.1002/clt2.12293. Clin Transl Allergy. 2023. PMID: 37876037 Free PMC article.
-
PLAUR splicing pattern in hereditary angioedema patients' monocytes and macrophages.Mol Biol Rep. 2023 Jun;50(6):4975-4982. doi: 10.1007/s11033-023-08391-8. Epub 2023 Apr 22. Mol Biol Rep. 2023. PMID: 37086298
-
Secreted uPAR isoform 2 (uPAR7b) is a novel direct target of miR-221.Oncotarget. 2015 Apr 10;6(10):8103-14. doi: 10.18632/oncotarget.3516. Oncotarget. 2015. PMID: 25797271 Free PMC article.
References
-
- Laufs S, Schumacher J, Allgayer H. Urokinase-receptor (u-PAR): an essential player in multiple games of cancer: a review on its role in tumor progression, invasion, metastasis, proliferation/dormancy, clinical outcome and minimal residual disease. Cell Cycle. 2006;5:1760–1771. - PubMed
-
- Mazar AP. Urokinase plasminogen activator receptor choreographs multiple ligand interactions: implications for tumor progression and therapy. Clin Cancer Res. 2008;14:5649–5655. - PubMed
-
- Luther T, Kotzsch M, Meye A, Langerholc T, Fussel S, Olbricht N, Albrecht S, Ockert D, Muehlenweg B, Friedrich K, et al. Identification of a novel urokinase receptor splice variant and its prognostic relevance in breast cancer. Thromb Haemost. 2003;89:705–717. - PubMed
-
- Collen D. The plasminogen (fibrinolytic) system. Thromb Haemost. 1999;82:259–270. - PubMed
-
- Blasi F, Carmeliet P. uPAR: a versatile signallingorchestrator. Nat Rev Mol Cell Biol. 2002;3:932–943. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

