Multiple histone modifications in euchromatin promote heterochromatin formation by redundant mechanisms in Saccharomyces cerevisiae
- PMID: 19638198
- PMCID: PMC2724485
- DOI: 10.1186/1471-2199-10-76
Multiple histone modifications in euchromatin promote heterochromatin formation by redundant mechanisms in Saccharomyces cerevisiae
Abstract
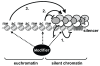
Background: Methylation of lysine 79 on histone H3 by Dot1 is required for maintenance of heterochromatin structure in yeast and humans. However, this histone modification occurs predominantly in euchromatin. Thus, Dot1 affects silencing by indirect mechanisms and does not act by the recruitment model commonly proposed for histone modifications. To better understand the role of H3K79 methylation gene silencing, we investigated the silencing function of Dot1 by genetic suppressor and enhancer analysis and examined the relationship between Dot1 and other global euchromatic histone modifiers.
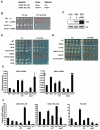
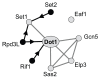
Result: We determined that loss of H3K79 methylation results in a partial silencing defect that could be bypassed by conditions that promote targeting of Sir proteins to heterochromatin. Furthermore, the silencing defect in strains lacking Dot1 was dependent on methylation of H3K4 by Set1 and histone acetylation by Gcn5, Elp3, and Sas2 in euchromatin. Our study shows that multiple histone modifications associated with euchromatin positively modulate the function of heterochromatin by distinct mechanisms. Genetic interactions between Set1 and Set2 suggested that the H3K36 methyltransferase Set2, unlike most other euchromatic modifiers, negatively affects gene silencing.
Conclusion: Our genetic dissection of Dot1's role in silencing in budding yeast showed that heterochromatin formation is modulated by multiple euchromatic histone modifiers that act by non-overlapping mechanisms. We discuss how euchromatic histone modifiers can make negative as well as positive contributions to gene silencing by competing with heterochromatin proteins within heterochromatin, within euchromatin, and at the boundary between euchromatin and heterochromatin.
Figures




References
-
- Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol. 2007;14:1008–1016. - PubMed
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
-
- Sims RJ, III, Reinberg D. Is there a code embedded in proteins that is based on post-translational modifications? Nat Rev Mol Cell Biol. 2008;9:815–820. - PubMed
-
- van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

