SUMOylation of the mitochondrial fission protein Drp1 occurs at multiple nonconsensus sites within the B domain and is linked to its activity cycle
- PMID: 19638400
- PMCID: PMC2775011
- DOI: 10.1096/fj.09-136630
SUMOylation of the mitochondrial fission protein Drp1 occurs at multiple nonconsensus sites within the B domain and is linked to its activity cycle
Abstract
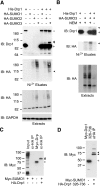
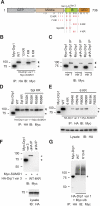
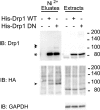
Dynamin-related protein (Drp) 1 is a key regulator of mitochondrial fission and is composed of GTP-binding, Middle, insert B, and C-terminal GTPase effector (GED) domains. Drp1 associates with mitochondrial fission sites and promotes membrane constriction through its intrinsic GTPase activity. The mechanisms that regulate Drp1 activity remain poorly understood but are likely to involve reversible post-translational modifications, such as conjugation of small ubiquitin-like modifier (SUMO) proteins. Through a detailed analysis, we find that Drp1 interacts with the SUMO-conjugating enzyme Ubc9 via multiple regions and demonstrate that Drp1 is a direct target of SUMO modification by all three SUMO isoforms. While Drp1 does not harbor consensus SUMOylation sequences, our analysis identified2 clusters of lysine residues within the B domain that serve as noncanonical conjugation sites. Although initial analysis indicates that mitochondrial recruitment of ectopically expressed Drp1 in response to staurosporine is unaffected by loss of SUMOylation, we find that Drp1 SUMOylation is enhanced in the context of the K38A mutation. This dominant-negative mutant, which is deficient in GTP binding and hydrolysis, does not associate with mitochondria and prevents normal mitochondrial fission. This finding suggests that SUMOylation of Drp1 is linked to its activity cycle and is influenced by Drp1 localization.
Figures


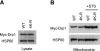
References
-
- Cerveny K L, Tamura Y, Zhang Z, Jensen R E, Sesaki H. Regulation of mitochondrial fusion and division. Trends Cell Biol. 2007;17:563–569. - PubMed
-
- Griparic L, van der Wel N N, Orozco I J, Peters P J, van der Bliek A M. Loss of the intermembrane space protein Mgm1/OPA1 induces swelling and localized constrictions along the lengths of mitochondria. J Biol Chem. 2004;279:18792–18798. - PubMed
-
- Santel A, Frank S, Gaume B, Herrler M, Youle R J, Fuller M T. Mitofusin-1 protein is a generally expressed mediator of mitochondrial fusion in mammalian cells. J Cell Sci. 2003;116:2763–2774. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

