Clueless, a conserved Drosophila gene required for mitochondrial subcellular localization, interacts genetically with parkin
- PMID: 19638420
- PMCID: PMC2737057
- DOI: 10.1242/dmm.002378
Clueless, a conserved Drosophila gene required for mitochondrial subcellular localization, interacts genetically with parkin
Abstract
Parkinson's disease has been linked to altered mitochondrial function. Mutations in parkin (park), the Drosophila ortholog of a human gene that is responsible for many familial cases of Parkinson's disease, shorten life span, abolish fertility and disrupt mitochondrial structure. However, the role played by Park in mitochondrial function remains unclear. Here, we describe a novel Drosophila gene, clueless (clu), which encodes a highly conserved tetratricopeptide repeat protein that is related closely to the CluA protein of Dictyostelium, Clu1 of Saccharomyces cerevisiae and to similar proteins in diverse metazoan eukaryotes from Arabidopsis to humans. Like its orthologs, loss of Drosophila clu causes mitochondria to cluster within cells. We find that strong clu mutations resemble park mutations in their effects on mitochondrial function and that the two genes interact genetically. Conversely, mitochondria in park homozygotes become highly clustered. We propose that Clu functions in a novel pathway that positions mitochondria within the cell based on their physiological state. Disruption of the Clu pathway may enhance oxidative damage, alter gene expression, cause mitochondria to cluster at microtubule plus ends, and lead eventually to mitochondrial failure.
Figures
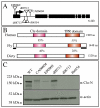


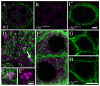
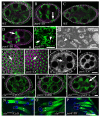

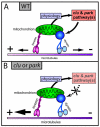
Similar articles
-
Clueless, a protein required for mitochondrial function, interacts with the PINK1-Parkin complex in Drosophila.Dis Model Mech. 2015 Jun;8(6):577-89. doi: 10.1242/dmm.019208. Epub 2015 Apr 20. Dis Model Mech. 2015. PMID: 26035866 Free PMC article.
-
Drosophila clueless is involved in Parkin-dependent mitophagy by promoting VCP-mediated Marf degradation.Hum Mol Genet. 2016 May 15;25(10):1946-1964. doi: 10.1093/hmg/ddw067. Epub 2016 Feb 29. Hum Mol Genet. 2016. PMID: 26931463 Free PMC article.
-
Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin.Nature. 2006 Jun 29;441(7097):1162-6. doi: 10.1038/nature04779. Epub 2006 May 3. Nature. 2006. PMID: 16672981
-
Drosophila clueless is highly expressed in larval neuroblasts, affects mitochondrial localization and suppresses mitochondrial oxidative damage.PLoS One. 2013;8(1):e54283. doi: 10.1371/journal.pone.0054283. Epub 2013 Jan 16. PLoS One. 2013. PMID: 23342118 Free PMC article.
-
Clueless forms dynamic, insulin-responsive bliss particles sensitive to stress.Dev Biol. 2020 Mar 15;459(2):149-160. doi: 10.1016/j.ydbio.2019.12.004. Epub 2019 Dec 16. Dev Biol. 2020. PMID: 31837288 Free PMC article.
Cited by
-
GRASP: A Multitasking Tether.Front Cell Dev Biol. 2016 Jan 26;4:1. doi: 10.3389/fcell.2016.00001. eCollection 2016. Front Cell Dev Biol. 2016. PMID: 26858948 Free PMC article. Review.
-
DRP1-dependent mitochondrial fission initiates follicle cell differentiation during Drosophila oogenesis.J Cell Biol. 2012 May 14;197(4):487-97. doi: 10.1083/jcb.201110058. J Cell Biol. 2012. PMID: 22584906 Free PMC article.
-
Clueless, a protein required for mitochondrial function, interacts with the PINK1-Parkin complex in Drosophila.Dis Model Mech. 2015 Jun;8(6):577-89. doi: 10.1242/dmm.019208. Epub 2015 Apr 20. Dis Model Mech. 2015. PMID: 26035866 Free PMC article.
-
Deciphering the evolution of composite-type GSKIP in mitochondria and Wnt signaling pathways.PLoS One. 2022 Jan 20;17(1):e0262138. doi: 10.1371/journal.pone.0262138. eCollection 2022. PLoS One. 2022. PMID: 35051222 Free PMC article.
-
Did mitophagy follow the origin of mitochondria?Autophagy. 2024 May;20(5):985-993. doi: 10.1080/15548627.2024.2307215. Epub 2024 Feb 15. Autophagy. 2024. PMID: 38361280 Free PMC article. Review.
References
-
- Abou-Sleiman PM, Muqit MM, Wood NW. (2006). Expanding insights of mitochondrial dysfunction in Parkinson’s disease. Nat Rev Neurosci. 7, 207–219 - PubMed
-
- Allen AK, Spradling AC. (2008). The Sf1-related nuclear hormone receptor Hr39 regulates Drosophila female reproductive tract development and function. Development 135, 311–321 - PubMed
-
- Betarbet R, Sherer TB, Greenamyre JT. (2005). Ubiquitin-proteasome system and Parkinson’s diseases. Exp Neurol. 191, S17–S27 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

