Whole-genome analyses reveal genetic instability of Acetobacter pasteurianus
- PMID: 19638423
- PMCID: PMC2761278
- DOI: 10.1093/nar/gkp612
Whole-genome analyses reveal genetic instability of Acetobacter pasteurianus
Abstract
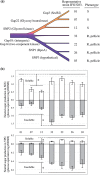
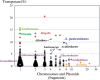
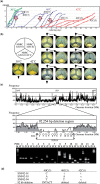
Acetobacter species have been used for brewing traditional vinegar and are known to have genetic instability. To clarify the mutability, Acetobacter pasteurianus NBRC 3283, which forms a multi-phenotype cell complex, was subjected to genome DNA sequencing. The genome analysis revealed that there are more than 280 transposons and five genes with hyper-mutable tandem repeats as common features in the genome consisting of a 2.9-Mb chromosome and six plasmids. There were three single nucleotide mutations and five transposon insertions in 32 isolates from the cell complex. The A. pasteurianus hyper-mutability was applied for breeding a temperature-resistant strain grown at an unviable high-temperature (42 degrees C). The genomic DNA sequence of a heritable mutant showing temperature resistance was analyzed by mutation mapping, illustrating that a 92-kb deletion and three single nucleotide mutations occurred in the genome during the adaptation. Alpha-proteobacteria including A. pasteurianus consists of many intracellular symbionts and parasites, and their genomes show increased evolution rates and intensive genome reduction. However, A. pasteurianus is assumed to be a free-living bacterium, it may have the potentiality to evolve to fit in natural niches of seasonal fruits and flowers with other organisms, such as yeasts and lactic acid bacteria.
Figures



References
-
- Lambert B, Kersters K, Gossele F, Swings J, De Ley J. Gluconobacters from honey bees. Antonie Van Leeuwenhoek. 1981;47:147–157. - PubMed
-
- Ryu JH, Kim SH, Lee HY, Bai JY, Nam YD, Bae JW, Lee DG, Shin SC, Ha EM, Lee WJ. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science. 2008;319:777–782. - PubMed
-
- Yamada Y, Yukphan P. Genera and species in acetic acid bacteria. Int. J. Food Microbiol. 2008;125:15–24. - PubMed
-
- Greenberg DE, Porcella SF, Stock F, Wong A, Conville PS, Murray PR, Holland SM, Zelazny AM. Granulibacter bethesdensis gen. nov., sp. nov., a distinctive pathogenic acetic acid bacterium in the family Acetobacteraceae. Int. J. Syst. Evol. Microbiol. 2006;56:2609–2616. - PubMed
-
- Adachi O, Moonmangmee D, Toyama H, Yamada M, Shinagawa E, Matsushita K. New developments in oxidative fermentation. Appl. Microbiol. Biotechnol. 2003;60:643–653. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

