Understanding modularity in molecular networks requires dynamics
- PMID: 19638611
- PMCID: PMC4243459
- DOI: 10.1126/scisignal.281pe44
Understanding modularity in molecular networks requires dynamics
Abstract
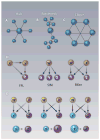
The era of genome sequencing has produced long lists of the molecular parts from which cellular machines are constructed. A fundamental goal in systems biology is to understand how cellular behavior emerges from the interaction in time and space of genetically encoded molecular parts, as well as nongenetically encoded small molecules. Networks provide a natural framework for the organization and quantitative representation of all the available data about molecular interactions. The structural and dynamic properties of molecular networks have been the subject of intense research. Despite major advances, bridging network structure to dynamics-and therefore to behavior-remains challenging. A key concept of modern engineering that recurs in the functional analysis of biological networks is modularity. Most approaches to molecular network analysis rely to some extent on the assumption that molecular networks are modular-that is, they are separable and can be studied to some degree in isolation. We describe recent advances in the analysis of modularity in biological networks, focusing on the increasing realization that a dynamic perspective is essential to grouping molecules into modules and determining their collective function.
Figures
References
-
- Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, Hao T, Rual JF, Dricot A, Vazquez A, Murray RR, Simon C, Tardivo L, Tam S, Svrzikapa N, Fan C, de Smet AS, Motyl A, Hudson ME, Park J, Xin X, Cusick ME, Moore T, Boone C, Snyder M, Roth FP, Barabasi AL, Tavernier J, Hill DE, Vidal M. High-Quality Binary Protein Interaction Map of the Yeast Interactome Network. Science. 2008;322:104–110. - PMC - PubMed
-
- Zhu XW, Gerstein M, Snyder M. Getting connected: analysis and principles of biological networks. Genes Dev. 2007;21:1010–1024. - PubMed
-
- Yu HY, Paccanaro A, Trifonov V, Gerstein M. Predicting interactions in protein networks by completing defective cliques. Bioinformatics. 2006;22:823–829. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

