Bile salts of vertebrates: structural variation and possible evolutionary significance
- PMID: 19638645
- PMCID: PMC2803226
- DOI: 10.1194/jlr.R000042
Bile salts of vertebrates: structural variation and possible evolutionary significance
Abstract
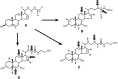
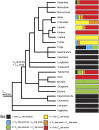
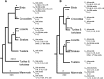
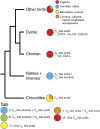
Biliary bile salt composition of 677 vertebrate species (103 fish, 130 reptiles, 271 birds, 173 mammals) was determined. Bile salts were of three types: C(27) bile alcohols, C(27) bile acids, or C(24) bile acids, with default hydroxylation at C-3 and C-7. C(27) bile alcohols dominated in early evolving fish and amphibians; C(27) bile acids, in reptiles and early evolving birds. C(24) bile acids were present in all vertebrate classes, often with C(27) alcohols or with C(27) acids, indicating two evolutionary pathways from C(27) bile alcohols to C(24) bile acids: a) a 'direct' pathway and b) an 'indirect' pathway with C(27) bile acids as intermediates. Hydroxylation at C-12 occurred in all orders and at C-16 in snakes and birds. Minor hydroxylation sites were C-1, C-2, C-5, C-6, and C-15. Side chain hydroxylation in C(27) bile salts occurred at C-22, C-24, C-25, and C-26, and in C(24) bile acids, at C-23 (snakes, birds, and pinnipeds). Unexpected was the presence of C(27) bile alcohols in four early evolving mammals. Bile salt composition showed significant variation between orders but not between families, genera, or species. Bile salt composition is a biochemical trait providing clues to evolutionary relationships, complementing anatomical and genetic analyses.
Figures
References
-
- Norlin M., Wikvall K. 2007. Enzymes in the conversion of cholesterol into bile acids. Curr. Mol. Med. 7: 199–218. - PubMed
-
- Russell D. W. 2003. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 72: 137–174. - PubMed
-
- Forman B. M., Goode E., Chen J., Oro A. E., Bradley D. J., Perlmann T., Noonan D. J., Burka L. T., McMorris T., Lamph W. W., et al. 1995. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell. 81: 687–693. - PubMed
-
- Makishima M., Okamato A. Y., Repa J. J., Tu H., Learned R. M., Luk A., Hull M. V., Lustig K. D., Mangelsdorf D. J., Shan B. 1999. Identification of a nuclear receptor for bile acids. Science. 284: 1362–1365. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

