Acyl-CoA dehydrogenases: Dynamic history of protein family evolution
- PMID: 19639238
- PMCID: PMC4136416
- DOI: 10.1007/s00239-009-9263-0
Acyl-CoA dehydrogenases: Dynamic history of protein family evolution
Abstract
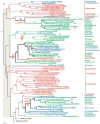
The acyl-CoA dehydrogenases (ACADs) are enzymes that catalyze the alpha,beta-dehydrogenation of acyl-CoA esters in fatty acid and amino acid catabolism. Eleven ACADs are now recognized in the sequenced human genome, and several homologs have been reported from bacteria, fungi, plants, and nematodes. We performed a systematic comparative genomic study, integrating homology searches with methods of phylogenetic reconstruction, to investigate the evolutionary history of this family. Sequence analyses indicate origin of the family in the common ancestor of Archaea, Bacteria, and Eukaryota, illustrating its essential role in the metabolism of early life. At least three ACADs were already present at that time: ancestral glutaryl-CoA dehydrogenase (GCD), isovaleryl-CoA dehydrogenase (IVD), and ACAD10/11. Two gene duplications were unique to the eukaryotic domain: one resulted in the VLCAD and ACAD9 paralogs and another in the ACAD10 and ACAD11 paralogs. The overall patchy distribution of specific ACADs across the tree of life is the result of dynamic evolution that includes numerous rounds of gene duplication and secondary losses, interdomain lateral gene transfer events, alteration of cellular localization, and evolution of novel proteins by domain acquisition. Our finding that eukaryotic ACAD species are more closely related to bacterial ACADs is consistent with endosymbiotic origin of ACADs in eukaryotes and further supported by the localization of all nine previously studied ACADs in mitochondria.
Figures





References
-
- Andresen BS, Bross P, Vianey-Saban C, Divry P, Zabot MT, Roe CR, Nada MA, Byskov A, Kruse TA, Neve S, Kristiansen K, Knudsen I, Corydon MJ, Gregersen N. Cloning and characterization of human very-long-chain acyl-CoA dehydrogenase cDNA, chromosomal assignment of the gene and identification in four patients of nine different mutations within the VLCAD gene. Hum Mol Genet. 1996;5:461–472. - PubMed
-
- Battaile K, Molin-Case J, Paschke R, Wang M, Bennett D, Vockley J, Kim JJP. Crystal structure of rat short chain acyl-CoA dehydrogenase complexed with acetoacetyl-CoA; comparison with other acyl-CoA dehydrogenases. J Biol Chem. 2002;277:12200–12207. - PubMed
-
- Battaile KP, Nguyen TV, Vockley J, Kim JJ. Structures of isobutyryl-CoA dehydrogenase and enzyme-product complex: comparison with isovaleryl- and short-chain acyl-CoA dehydrogenases. J Biol Chem. 2004;279:16526–16534. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

