Mammalian Par3 regulates progenitor cell asymmetric division via notch signaling in the developing neocortex
- PMID: 19640478
- PMCID: PMC2736606
- DOI: 10.1016/j.neuron.2009.07.004
Mammalian Par3 regulates progenitor cell asymmetric division via notch signaling in the developing neocortex
Abstract
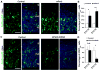
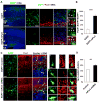
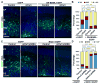
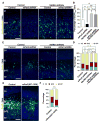
Asymmetric cell division of radial glial progenitors produces neurons while allowing self-renewal; however, little is known about the mechanism that generates asymmetry in daughter cell fate specification. Here, we found that mammalian partition defective protein 3 (mPar3), a key cell polarity determinant, exhibits dynamic distribution in radial glial progenitors. While it is enriched at the lateral membrane domain in the ventricular endfeet during interphase, mPar3 becomes dispersed and shows asymmetric localization as cell cycle progresses. Either removal or ectopic expression of mPar3 prevents radial glial progenitors from dividing asymmetrically yet generates different outcomes in daughter cell fate specification. Furthermore, the expression level of mPar3 affects Notch signaling, and manipulations of Notch signaling or Numb expression suppress mPar3 regulation of radial glial cell division and daughter cell fate specification. These results reveal a critical molecular pathway underlying asymmetric cell division of radial glial progenitors in the mammalian neocortex.
Figures
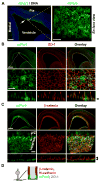
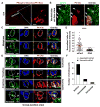
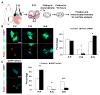
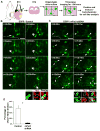
References
-
- Aaku-Saraste E, Hellwig A, Huttner WB. Loss of occludin and functional tight junctions, but not ZO-1, during neural tube closure--remodeling of the neuroepithelium prior to neurogenesis. Dev Biol. 1996;180:664–679. - PubMed
-
- Anthony TE, Klein C, Fishell G, Heintz N. Radial glia serve as neuronal progenitors in all regions of the central nervous system. Neuron. 2004;41:881–890. - PubMed
-
- Astrom KE, Webster HD. The early development of the neopallial wall and area choroidea in fetal rats. A light and electron microscopic study. Adv Anat Embryol Cell Biol. 1991;123:1–76. - PubMed
-
- Buchman JJ, Tsai LH. Spindle regulation in neural precursors of flies and mammals. Nat Rev Neurosci. 2007;8:89–100. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

