Mechanism of cyclizing NAD to cyclic ADP-ribose by ADP-ribosyl cyclase and CD38
- PMID: 19640843
- PMCID: PMC2785691
- DOI: 10.1074/jbc.M109.030965
Mechanism of cyclizing NAD to cyclic ADP-ribose by ADP-ribosyl cyclase and CD38
Abstract
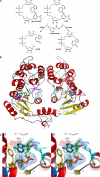
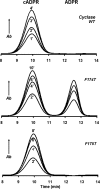
Mammalian CD38 and its Aplysia homolog, ADP-ribosyl cyclase (cyclase), are two prominent enzymes that catalyze the synthesis and hydrolysis of cyclic ADP-ribose (cADPR), a Ca(2+) messenger molecule responsible for regulating a wide range of cellular functions. Although both use NAD as a substrate, the cyclase produces cADPR, whereas CD38 produces mainly ADP-ribose (ADPR). To elucidate the catalytic differences and the mechanism of cyclizing NAD, the crystal structure of a stable complex of the cyclase with an NAD analog, ribosyl-2'F-2'deoxynicotinamide adenine dinucleotide (ribo-2'-F-NAD), was determined. The results show that the analog was a substrate of the cyclase and that during the reaction, the nicotinamide group was released and a stable intermediate was formed. The terminal ribosyl unit at one end of the intermediate formed a close linkage with the catalytic residue (Glu-179), whereas the adenine ring at the other end stacked closely with Phe-174, suggesting that the latter residue is likely to be responsible for folding the linear substrate so that the two ends can be cyclized. Mutating Phe-174 indeed reduced cADPR production but enhanced ADPR production, converting the cyclase to be more CD38-like. Changing the equivalent residue in CD38, Thr-221 to Phe, correspondingly enhanced cADPR production, and the double mutation, Thr-221 to Phe and Glu-146 to Ala, effectively converted CD38 to a cyclase. This study provides the first detailed evidence of the cyclization process and demonstrates the feasibility of engineering the reactivity of the enzymes by mutation, setting the stage for the development of tools to manipulate cADPR metabolism in vivo.
Figures


Similar articles
-
Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase.J Biol Chem. 2009 Oct 2;284(40):27637-45. doi: 10.1074/jbc.M109.031005. Epub 2009 Jul 28. J Biol Chem. 2009. PMID: 19640846 Free PMC article.
-
Porcine CD38 exhibits prominent secondary NAD(+) cyclase activity.Protein Sci. 2016 Mar;25(3):650-61. doi: 10.1002/pro.2859. Epub 2016 Jan 12. Protein Sci. 2016. PMID: 26660500 Free PMC article.
-
Structural basis for formation and hydrolysis of the calcium messenger cyclic ADP-ribose by human CD38.J Biol Chem. 2007 Feb 23;282(8):5853-61. doi: 10.1074/jbc.M609093200. Epub 2006 Dec 20. J Biol Chem. 2007. PMID: 17182614
-
Resolving the topological enigma in Ca2+ signaling by cyclic ADP-ribose and NAADP.J Biol Chem. 2019 Dec 27;294(52):19831-19843. doi: 10.1074/jbc.REV119.009635. Epub 2019 Oct 31. J Biol Chem. 2019. PMID: 31672920 Free PMC article. Review.
-
Paracrine ADP Ribosyl Cyclase-Mediated Regulation of Biological Processes.Cells. 2022 Aug 24;11(17):2637. doi: 10.3390/cells11172637. Cells. 2022. PMID: 36078044 Free PMC article. Review.
Cited by
-
Selective targeting of CD38 hydrolase and cyclase activity as an approach to immunostimulation.RSC Adv. 2021 Oct 11;11(53):33260-33270. doi: 10.1039/d1ra06266b. eCollection 2021 Oct 8. RSC Adv. 2021. PMID: 35497564 Free PMC article.
-
CD38 Structure-Based Inhibitor Design Using the N1-Cyclic Inosine 5'-Diphosphate Ribose Template.PLoS One. 2013 Jun 19;8(6):e66247. doi: 10.1371/journal.pone.0066247. Print 2013. PLoS One. 2013. PMID: 23840430 Free PMC article.
-
Daratumumab and Nanobody-Based Heavy Chain Antibodies Inhibit the ADPR Cyclase but not the NAD+ Hydrolase Activity of CD38-Expressing Multiple Myeloma Cells.Cancers (Basel). 2020 Dec 30;13(1):76. doi: 10.3390/cancers13010076. Cancers (Basel). 2020. PMID: 33396591 Free PMC article.
-
CD38 deficiency suppresses adipogenesis and lipogenesis in adipose tissues through activating Sirt1/PPARγ signaling pathway.J Cell Mol Med. 2018 Jan;22(1):101-110. doi: 10.1111/jcmm.13297. Epub 2017 Aug 16. J Cell Mol Med. 2018. PMID: 28816006 Free PMC article.
-
Snake venom NAD glycohydrolases: primary structures, genomic location, and gene structure.PeerJ. 2019 Feb 6;7:e6154. doi: 10.7717/peerj.6154. eCollection 2019. PeerJ. 2019. PMID: 30755823 Free PMC article.
References
-
- Clapper D. L., Walseth T. F., Dargie P. J., Lee H. C. (1987) J. Biol. Chem. 262, 9561–9568 - PubMed
-
- Lee H. C., Walseth T. F., Bratt G. T., Hayes R. N., Clapper D. L. (1989) J. Biol. Chem. 264, 1608–1615 - PubMed
-
- Lam C. M., Yeung P. K., Lee H. C., Wong J. T. (2009) Cell Calcium 45, 346–357 - PubMed
-
- Jin D., Liu H. X., Hirai H., Torashima T., Nagai T., Lopatina O., Shnayder N. A., Yamada K., Noda M., Seike T., Fujita K., Takasawa S., Yokoyama S., Koizumi K., Shiraishi Y., Tanaka S., Hashii M., Yoshihara T., Higashida K., Islam M. S., Yamada N., Hayashi K., Noguchi N., Kato I., Okamoto H., Matsushima A., Salmina A., Munesue T., Shimizu N., Mochida S., Asano M., Higashida H. (2007) Nature 446, 41–45 - PubMed
-
- Lee H. C. (2002) Cyclic ADP-ribose and NAADP: Structures, Metabolism and Functions, Kluwer Academic Publishers, Dordrecht, The Netherlands
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

