Crystal structure of a homolog of mammalian serine racemase from Schizosaccharomyces pombe
- PMID: 19640845
- PMCID: PMC2757995
- DOI: 10.1074/jbc.M109.010470
Crystal structure of a homolog of mammalian serine racemase from Schizosaccharomyces pombe
Abstract
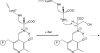
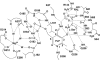
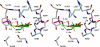
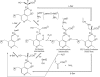
D-serine is an endogenous coagonist for the N-methyl-D-aspartate receptor and is involved in excitatory neurotransmission in the brain. Mammalian pyridoxal 5'-phosphate-dependent serine racemase, which is localized in the mammalian brain, catalyzes the racemization of L-serine to yield D-serine and vice versa. The enzyme also catalyzes the dehydration of D- and L-serine. Both reactions are enhanced by Mg.ATP in vivo. We have determined the structures of the following three forms of the mammalian enzyme homolog from Schizosaccharomyces pombe: the wild-type enzyme, the wild-type enzyme in the complex with an ATP analog, and the modified enzyme in the complex with serine at 1.7, 1.9, and 2.2 A resolution, respectively. On binding of the substrate, the small domain rotates toward the large domain to close the active site. The ATP binding site was identified at the domain and the subunit interface. Computer graphics models of the wild-type enzyme complexed with L-serine and D-serine provided an insight into the catalytic mechanisms of both reactions. Lys-57 and Ser-82 located on the protein and solvent sides, respectively, with respect to the cofactor plane, are acid-base catalysts that shuttle protons to the substrate. The modified enzyme, which has a unique "lysino-D-alanyl" residue at the active site, also exhibits catalytic activities. The crystal-soaking experiment showed that the substrate serine was actually trapped in the active site of the modified enzyme, suggesting that the lysino-D-alanyl residue acts as a catalytic base in the same manner as inherent Lys-57 of the wild-type enzyme.
Figures



Similar articles
-
Serine racemase with catalytically active lysinoalanyl residue.J Biochem. 2009 Apr;145(4):421-4. doi: 10.1093/jb/mvp010. Epub 2009 Jan 20. J Biochem. 2009. PMID: 19155267
-
Metal ion dependency of serine racemase from Dictyostelium discoideum.Amino Acids. 2012 Oct;43(4):1567-76. doi: 10.1007/s00726-012-1232-z. Epub 2012 Feb 5. Amino Acids. 2012. PMID: 22311068
-
ATP binding to human serine racemase is cooperative and modulated by glycine.FEBS J. 2013 Nov;280(22):5853-63. doi: 10.1111/febs.12510. Epub 2013 Sep 25. FEBS J. 2013. PMID: 23992455
-
D-amino acids in the brain: the biochemistry of brain serine racemase.FEBS J. 2008 Jul;275(14):3538-45. doi: 10.1111/j.1742-4658.2008.06517.x. FEBS J. 2008. PMID: 18564178 Review.
-
Serine racemase: a key player in neuron activity and in neuropathologies.Front Biosci (Landmark Ed). 2013 Jun 1;18(3):1112-28. doi: 10.2741/4167. Front Biosci (Landmark Ed). 2013. PMID: 23747871 Review.
Cited by
-
Crystal structure of a zinc-dependent D-serine dehydratase from chicken kidney.J Biol Chem. 2011 Aug 5;286(31):27548-58. doi: 10.1074/jbc.M110.201160. Epub 2011 Jun 15. J Biol Chem. 2011. PMID: 21676877 Free PMC article.
-
Serine Racemase Expression by Striatal Neurons.Cell Mol Neurobiol. 2022 Jan;42(1):279-289. doi: 10.1007/s10571-020-00880-9. Epub 2020 May 22. Cell Mol Neurobiol. 2022. PMID: 32445040 Free PMC article.
-
D-Serine Metabolism and Its Importance in Development of Dictyostelium discoideum.Front Microbiol. 2018 Apr 24;9:784. doi: 10.3389/fmicb.2018.00784. eCollection 2018. Front Microbiol. 2018. PMID: 29740415 Free PMC article.
-
Human Serine Racemase: Key Residues/Active Site Motifs and Their Relation to Enzyme Function.Front Mol Biosci. 2019 Mar 13;6:8. doi: 10.3389/fmolb.2019.00008. eCollection 2019. Front Mol Biosci. 2019. PMID: 30918891 Free PMC article. Review.
-
Involvement of C-terminal amino acids of a hyperthermophilic serine racemase in its thermostability.Extremophiles. 2018 Jan;22(1):99-107. doi: 10.1007/s00792-017-0980-9. Epub 2017 Nov 9. Extremophiles. 2018. PMID: 29124361
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

