Quantitative phosphokinome analysis of the Met pathway activated by the invasin internalin B from Listeria monocytogenes
- PMID: 19640851
- PMCID: PMC2816026
- DOI: 10.1074/mcp.M800521-MCP200
Quantitative phosphokinome analysis of the Met pathway activated by the invasin internalin B from Listeria monocytogenes
Abstract
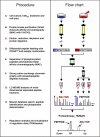
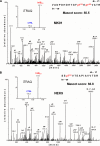
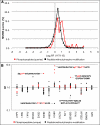
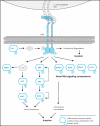
Stimulated by its physiological ligand, hepatocyte growth factor, the transmembrane receptor tyrosine kinase Met activates a signaling machinery that leads to mitogenic, motogenic, and morphogenic responses. Remarkably, the food-borne human pathogen Listeria monocytogenes also promotes autophosphorylation of Met through its virulence factor internalin B (InlB) and subsequently exploits Met signaling to induce phagocytosis into a broad range of host cells. Although the interaction between InlB and Met has been studied in detail, the signaling specificity of components involved in InlB-triggered cellular responses remains poorly characterized. The analysis of regulated phosphorylation events on protein kinases is therefore of particular relevance, although this could not as yet be characterized systematically by proteomics. Here, we implemented a new pyridopyrimidine-based strategy that enabled the efficient capture of a considerable subset of the human kinome in a robust one-step affinity chromatographic procedure. Additionally, and to gain functional insights into the InlB/Met-induced bacterial invasion process, a quantitative survey of the phosphorylation pattern of these protein kinases was accomplished. In total, the experimental design of this study comprises affinity chromatographic procedures for the systematic enrichment of kinases, as well as phosphopeptides; the quantification of all peptides based on the iTRAQ reporter system; and a rational statistical strategy to evaluate the quality of phosphosite regulations. With this improved chemical proteomics strategy, we determined and relatively quantified 143 phosphorylation sites detected on 94 human protein kinases. Interestingly, InlB-mediated signaling shows striking similarities compared with the natural ligand hepatocyte growth factor that was intensively studied in the past. In addition, this systematic approach suggests a new subset of protein kinases including Nek9, which are differentially phosphorylated after short time (4-min) treatment of cells with the Met-activating InlB(321). Thus, this quantitative phosphokinome study suggests a general, hypothesis-free concept for the detection of dynamically regulated protein kinases as novel signaling components involved in host-pathogen interactions.
Figures



Similar articles
-
InIB-dependent internalization of Listeria is mediated by the Met receptor tyrosine kinase.Cell. 2000 Oct 27;103(3):501-10. doi: 10.1016/s0092-8674(00)00141-0. Cell. 2000. PMID: 11081636
-
MET-activating Residues in the B-repeat of the Listeria monocytogenes Invasion Protein InlB.J Biol Chem. 2016 Dec 2;291(49):25567-25577. doi: 10.1074/jbc.M116.746685. Epub 2016 Oct 27. J Biol Chem. 2016. PMID: 27789707 Free PMC article.
-
Structural basis of VHH-mediated neutralization of the food-borne pathogen Listeria monocytogenes.J Biol Chem. 2018 Aug 31;293(35):13626-13635. doi: 10.1074/jbc.RA118.003888. Epub 2018 Jul 5. J Biol Chem. 2018. PMID: 29976754 Free PMC article.
-
Structural basis of MET receptor dimerization by the bacterial invasion protein InlB and the HGF/SF splice variant NK1.Biochim Biophys Acta. 2013 Oct;1834(10):2195-204. doi: 10.1016/j.bbapap.2012.10.012. Epub 2012 Oct 31. Biochim Biophys Acta. 2013. PMID: 23123275 Review.
-
InlB, a surface protein of Listeria monocytogenes that behaves as an invasin and a growth factor.J Cell Sci. 2002 Sep 1;115(Pt 17):3357-67. doi: 10.1242/jcs.115.17.3357. J Cell Sci. 2002. PMID: 12154067 Review.
Cited by
-
Host Serine/Threonine Kinases mTOR and Protein Kinase C-α Promote InlB-Mediated Entry of Listeria monocytogenes.Infect Immun. 2017 Jun 20;85(7):e00087-17. doi: 10.1128/IAI.00087-17. Print 2017 Jul. Infect Immun. 2017. PMID: 28461391 Free PMC article.
-
Kinome analysis of receptor-induced phosphorylation in human natural killer cells.PLoS One. 2012;7(1):e29672. doi: 10.1371/journal.pone.0029672. Epub 2012 Jan 4. PLoS One. 2012. PMID: 22238634 Free PMC article.
-
Recognition of host proteins by Helicobacter cysteine-rich protein C.Curr Microbiol. 2011 Sep;63(3):239-49. doi: 10.1007/s00284-011-9969-2. Epub 2011 Jul 7. Curr Microbiol. 2011. PMID: 21735226
-
Chronic Toxoplasma infection is associated with distinct alterations in the synaptic protein composition.J Neuroinflammation. 2018 Aug 1;15(1):216. doi: 10.1186/s12974-018-1242-1. J Neuroinflammation. 2018. PMID: 30068357 Free PMC article.
-
Analysis of differentially expressed proteins in Yersinia enterocolitica-infected HeLa cells.Biochim Biophys Acta. 2016 May;1864(5):562-9. doi: 10.1016/j.bbapap.2016.02.004. Epub 2016 Feb 5. Biochim Biophys Acta. 2016. PMID: 26854600 Free PMC article.
References
-
- Lecuit M., Vandormael-Pournin S., Lefort J., Huerre M., Gounon P., Dupuy C., Babinet C., Cossart P. ( 2001) A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science 292, 1722– 1725 - PubMed
-
- Mengaud J., Ohayon H., Gounon P., Mege R.-M., Cossart P. ( 1996) E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell 84, 923– 932 - PubMed
-
- Dramsi S., Biswas I., Maguin E., Braun L., Mastroeni P., Cossart P. ( 1995) Entry of Listeria monocytogenes into hepatocytes requires expression of inIB, a surface protein of the internalin multigene family. Mol. Microbiol 16, 251– 261 - PubMed
-
- Parida S. K., Domann E., Rohde M., Müller S., Darji A., Hain T., Wehland J., Chakraborty T. ( 1998) Internalin B is essential for adhesion and mediates the invasion of Listeria monocytogenes into human endothelial cells. Mol. Microbiol 28, 81– 93 - PubMed
-
- Trost M., Wehmhöner D., Kärst U., Dieterich G., Wehland J., Jänsch L. ( 2005) Comparative proteome analysis of secretory proteins from pathogenic and nonpathogenic Listeria species. Proteomics 5, 1544– 1557 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

