The renal H+-K+-ATPases: physiology, regulation, and structure
- PMID: 19640897
- PMCID: PMC2806118
- DOI: 10.1152/ajprenal.90723.2008
The renal H+-K+-ATPases: physiology, regulation, and structure
Abstract
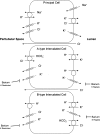
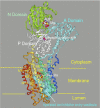
The H(+)-K(+)-ATPases are ion pumps that use the energy of ATP hydrolysis to transport protons (H(+)) in exchange for potassium ions (K(+)). These enzymes consist of a catalytic alpha-subunit and a regulatory beta-subunit. There are two catalytic subunits present in the kidney, the gastric or HKalpha(1) isoform and the colonic or HKalpha(2) isoform. In this review we discuss new information on the physiological function, regulation, and structure of the renal H(+)-K(+)-ATPases. Evaluation of enzymatic functions along the nephron and collecting duct and studies in HKalpha(1) and HKalpha(2) knockout mice suggest that the H(+)-K(+)-ATPases may function to transport ions other than protons and potassium. These reports and recent studies in mice lacking both HKalpha(1) and HKalpha(2) suggest important roles for the renal H(+)-K(+)-ATPases in acid/base balance as well as potassium and sodium homeostasis. Molecular modeling studies based on the crystal structure of a related enzyme have made it possible to evaluate the structures of HKalpha(1) and HKalpha(2) and provide a means to study the specific cation transport properties of H(+)-K(+)-ATPases. Studies to characterize the cation specificity of these enzymes under different physiological conditions are necessary to fully understand the role of the H(+)-K(+) ATPases in renal physiology.
Figures
Similar articles
-
Mineralocorticoids stimulate the activity and expression of renal H+,K+-ATPases.J Am Soc Nephrol. 2011 Jan;22(1):49-58. doi: 10.1681/ASN.2010030311. Epub 2010 Dec 16. J Am Soc Nephrol. 2011. PMID: 21164026 Free PMC article.
-
[Molecular and functional diversity of NA,K-ATPase and renal H,K-ATPases].Nephrologie. 1996;17(7):401-8. Nephrologie. 1996. PMID: 9019667 Review. French.
-
Impaired acid secretion in cortical collecting duct intercalated cells from H-K-ATPase-deficient mice: role of HKalpha isoforms.Am J Physiol Renal Physiol. 2008 Mar;294(3):F621-7. doi: 10.1152/ajprenal.00412.2007. Epub 2007 Dec 5. Am J Physiol Renal Physiol. 2008. PMID: 18057185
-
Pharmacological profiles of the murine gastric and colonic H,K-ATPases.Biochim Biophys Acta. 2010 Sep;1800(9):906-11. doi: 10.1016/j.bbagen.2010.05.002. Epub 2010 May 25. Biochim Biophys Acta. 2010. PMID: 20594946 Free PMC article.
-
[Renal K-ATPases: structure, function and dysfunction].Nephrologie. 1999;20(6):319-27. Nephrologie. 1999. PMID: 10592934 Review. French.
Cited by
-
Intracellular cAMP Sensor EPAC: Physiology, Pathophysiology, and Therapeutics Development.Physiol Rev. 2018 Apr 1;98(2):919-1053. doi: 10.1152/physrev.00025.2017. Physiol Rev. 2018. PMID: 29537337 Free PMC article. Review.
-
An evaluation of the protective effect of esomeprazole in an experimental model of renal ischemia-reperfusion.Int Urol Nephrol. 2018 Feb;50(2):217-223. doi: 10.1007/s11255-017-1775-8. Epub 2017 Dec 26. Int Urol Nephrol. 2018. PMID: 29280047
-
TRPV4 expression in the renal tubule is necessary for maintaining whole body K+ homeostasis.Am J Physiol Renal Physiol. 2023 Jun 1;324(6):F603-F616. doi: 10.1152/ajprenal.00278.2022. Epub 2023 May 4. Am J Physiol Renal Physiol. 2023. PMID: 37141145 Free PMC article.
-
A new look at electrolyte transport in the distal tubule.Annu Rev Physiol. 2012;74:325-49. doi: 10.1146/annurev-physiol-020911-153225. Epub 2011 Sep 2. Annu Rev Physiol. 2012. PMID: 21888509 Free PMC article. Review.
-
Role of epithelial sodium channel-related inflammation in human diseases.Front Immunol. 2023 Jul 25;14:1178410. doi: 10.3389/fimmu.2023.1178410. eCollection 2023. Front Immunol. 2023. PMID: 37559717 Free PMC article. Review.
References
-
- Ahn KY, Kone BC. Expression and cellular localization of mRNA encoding the “gastric” isoform of the H+-K+-ATPase in rat kidney. Am J Physiol Renal Fluid Electrolyte Physiol 268: F99–F109, 1995 - PubMed
-
- Ahn KY, Park KY, Kim KK, Kone BC. Chronic hypokalemia enhances expression of the H+-K+-ATPase α2-subunit gene in renal medulla. Am J Physiol Renal Fluid Electrolyte Physiol 271: F314–F321, 1996 - PubMed
-
- Ahn KY, Turner PB, Madsen KM, Kone BC. Effects of chronic hypokalemia on renal expression of the “gastric” H+-K+-ATPase α-subunit gene. Am J Physiol Renal Fluid Electrolyte Physiol 270: F557–F566, 1996 - PubMed
-
- Armitage FE, Wingo CS. Luminal acidification in the K-replete OMCDi: contributions of H-K-ATPase and bafilomycin-A1-sensitive H-ATPase. Am J Physiol Renal Fluid Electrolyte Physiol 267: F450–F458, 1994 - PubMed
-
- Asano S, Yoshida A, Yashiro H, Kobayashi Y, Morisato A, Ogawa H, Takeguchi N, Morii M. The cavity structure for docking the K+-competitive inhibitors in the gastric proton pump. J Biol Chem 279: 13968–13975, 2004 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

