A genetic variant of hepatitis B virus divergent from known human and ape genotypes isolated from a Japanese patient and provisionally assigned to new genotype J
- PMID: 19640977
- PMCID: PMC2753143
- DOI: 10.1128/JVI.00462-09
A genetic variant of hepatitis B virus divergent from known human and ape genotypes isolated from a Japanese patient and provisionally assigned to new genotype J
Abstract
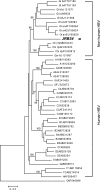
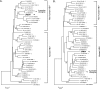
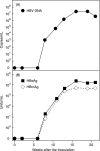
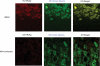
Hepatitis B virus (HBV) of a novel genotype (J) was recovered from an 88-year-old Japanese patient with hepatocellular carcinoma who had a history of residing in Borneo during the World War II. It was divergent from eight human (A to H) and four ape (chimpanzee, gorilla, gibbon, and orangutan) HBV genotypes, as well as from a recently proposed ninth human genotype I, by 9.9 to 16.5% of the entire genomic sequence and did not have evidence of recombination with any of the nine human genotypes and four nonhuman genotypes. Based on a comparison of the entire nucleotide sequence against 1,440 HBV isolates reported, HBV/J was nearest to the gibbon and orangutan genotypes (mean divergences of 10.9 and 10.7%, respectively). Based on a comparison of four open reading frames, HBV/J was closer to gibbon/orangutan genotypes than to human genotypes in the P and large S genes and closest to Australian aboriginal strains (HBV/C4) and orangutan-derived strains in the S gene, whereas it was closer to human than ape genotypes in the C gene. HBV/J shared a deletion of 33 nucleotides at the start of preS1 region with C4 and gibbon genotypes, had an S-gene sequence similar to that of C4, and expressed the ayw subtype. Efficient infection, replication, and antigen expression by HBV/J were experimentally established in two chimeric mice with the liver repopulated for human hepatocytes. The HBV DNA sequence recovered from infected mice was identical to that in the inoculum. Since HBV/J is positioned phylogenetically in between human and ape genotypes, it may help to trace the origin of HBV and merits further epidemiological surveys.
Figures


References
-
- Allen, M. I., M. Deslauriers, C. W. Andrews, G. A. Tipples, K. A. Walters, D. L. Tyrrell, N. Brown, L. D. Condreay, et al. 1998. Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Hepatology 27:1670-1677. - PubMed
-
- Arauz-Ruiz, P., H. Norder, B. H. Robertson, and L. O. Magnius. 2002. Genotype H: a new Amerindian genotype of hepatitis B virus revealed in Central America. J. Gen. Virol. 83:2059-2073. - PubMed
-
- Bollyky, P. L., and E. C. Holmes. 1999. Reconstructing the complex evolutionary history of hepatitis B virus. J. Mol. Evol. 49:130-141. - PubMed
-
- Carman, W. F., M. R. Jacyna, S. Hadziyannis, P. Karayiannis, M. J. McGarvey, A. Makris, and H. C. Thomas. 1989. Mutation preventing formation of hepatitis B e antigen in patients with chronic hepatitis B infection. Lancet ii:588-591. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

